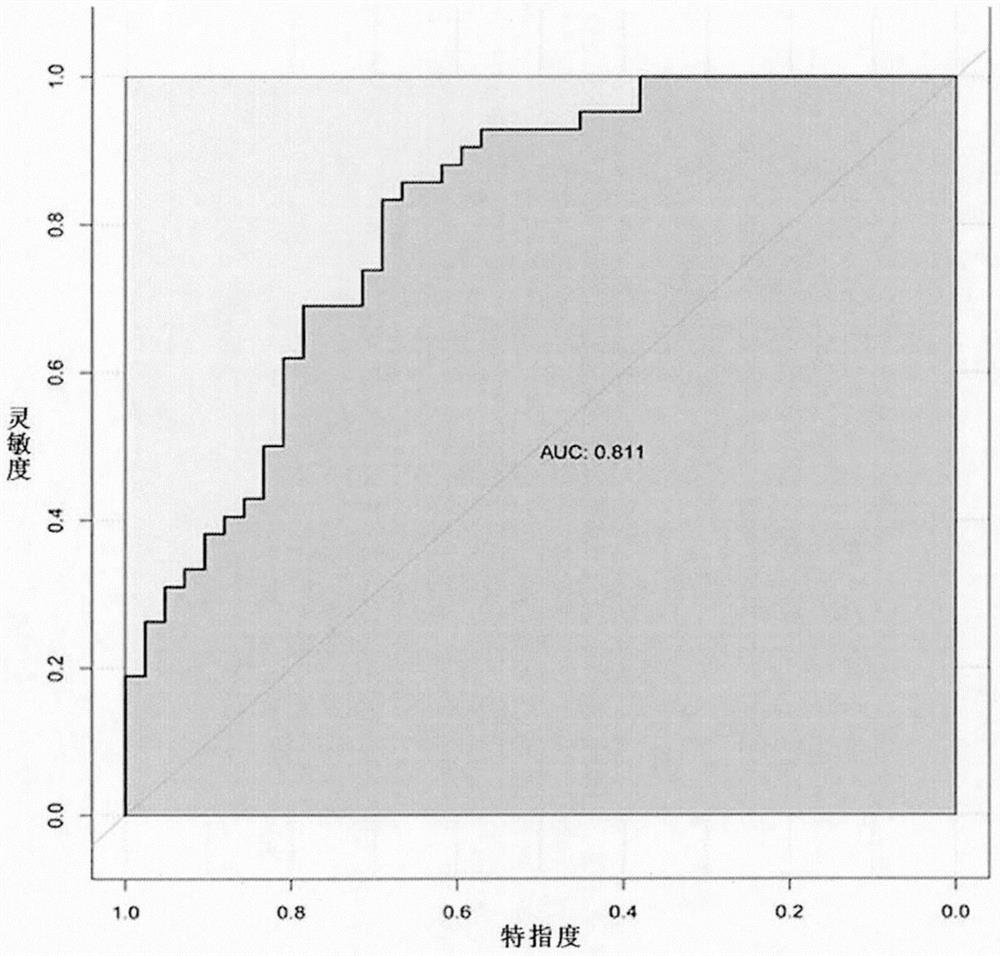
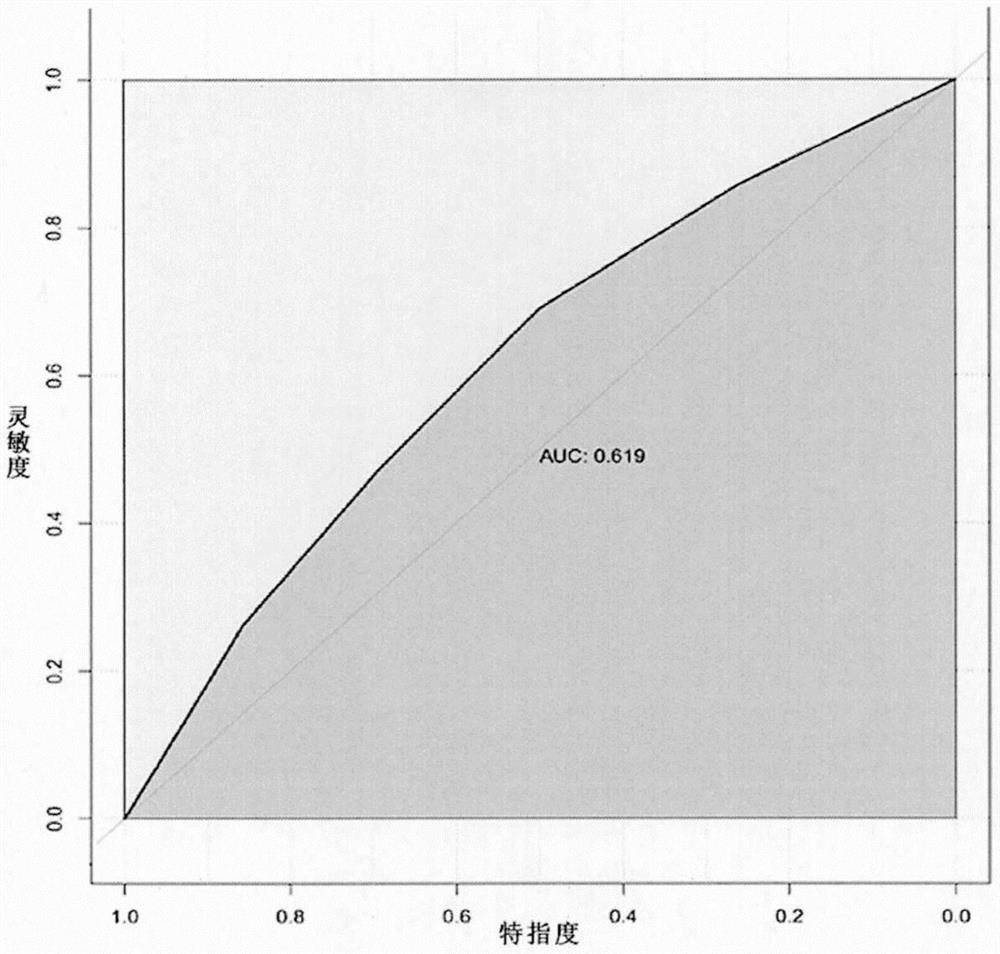
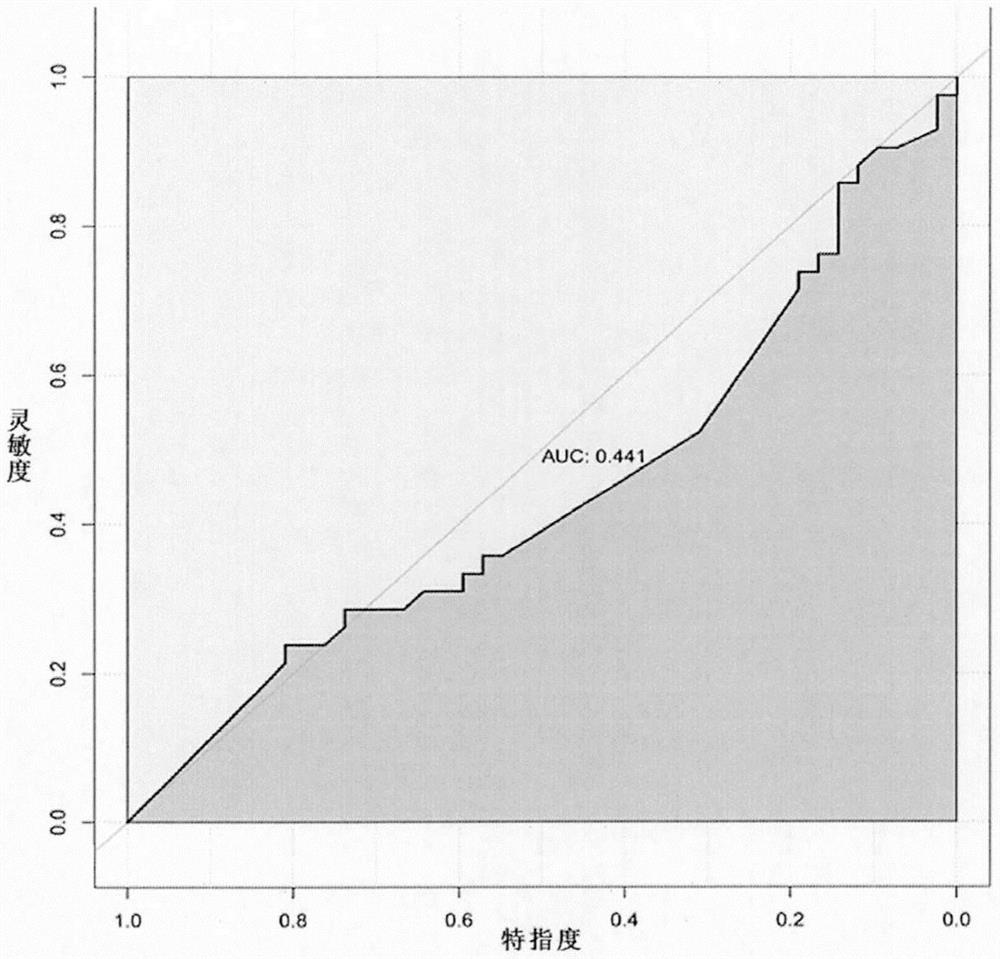
Small sample-based genotype and phenotype association analysis method in multi-omics data
An omics data, small sample technology, applied in the field of genotype and phenotype correlation research in multi-omics data based on small samples, can solve the difficulty of obtaining clinical data, and cannot meet the needs of multi-omics data fusion method data, SNP features are large and other problems, to achieve the effect of improving the prediction accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific example
[0057] 1. Data source and preprocessing
[0058] To verify the effectiveness of the method, the present invention uses two sets of data derived from the GEO database (Gene Expression Omnibus database, https: / / www.ncbi.nlm.nih.gov / geo / ) to verify. GSE33356 is studying lung adenocarcinoma. It includes lung cancer patients and their adjacent normal tissues, which are harvested from the patients. Lung tumors and normal specimens from 84 non-smoking female patients with adenocarcinoma were analyzed using Affymetrix SNP 6.0 and Affymetrix U133plus2.0 chips. GSE114269 is the data comparing myeloid breast cancer (MBC) and non-myeloid basal-like breast cancer (non-MBC BLC), with a sample size of 48. The main reason for choosing these two sets of data for experiments is to illustrate that the method of the present invention can be widely applied to such genotype and phenotype classification problems based on small sample multi-attributes.
[0059] The protein network data comes from ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com