Characterization and application of novel high-temperature Argonaute protein
A technology of reaction and reaction system, applied in the field of molecular biology and biology, can solve the problems of cumbersome steps and high detection cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0229] Embodiment 1: Obtaining of TeAgo gene sequence
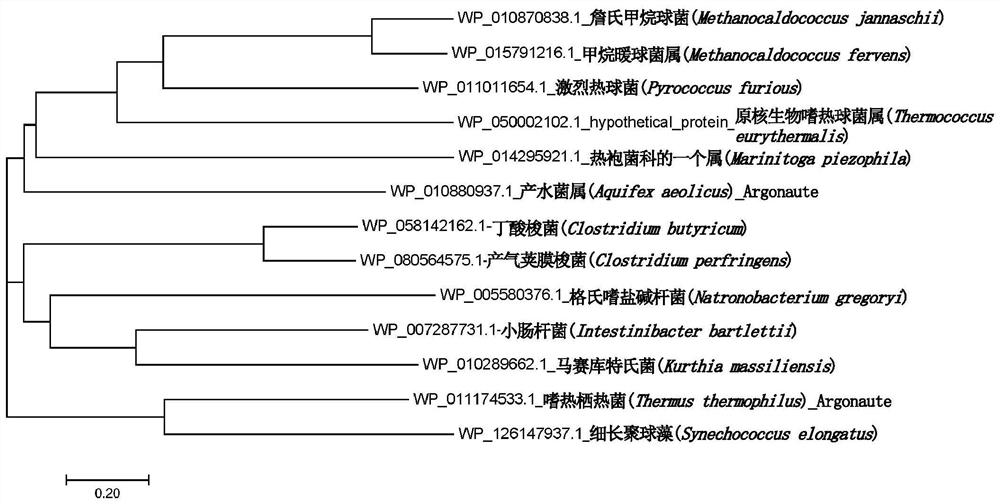
[0230]In the database, the amino acid sequence of known PfAgo is searched for similarity, and some amino acid sequences with high sequence consistency are selected, analyzed by MEGA software, and a homologous evolution tree is constructed, and TeAgo is selected as a candidate enzyme, wherein TeAgo and The sequence similarity of the characterized PfAgo reaches 33.02%. The amino acid sequence of TeAgo (WP_050002102.1) and the corresponding gene sequence (NZ_CP008887.1) encoding the protein were obtained. After the gene sequence was synthesized by codon optimization, it was cloned into pET28a expression vector.
Embodiment 2
[0231] Example 2: TeAgo protein heterologous expression and purification
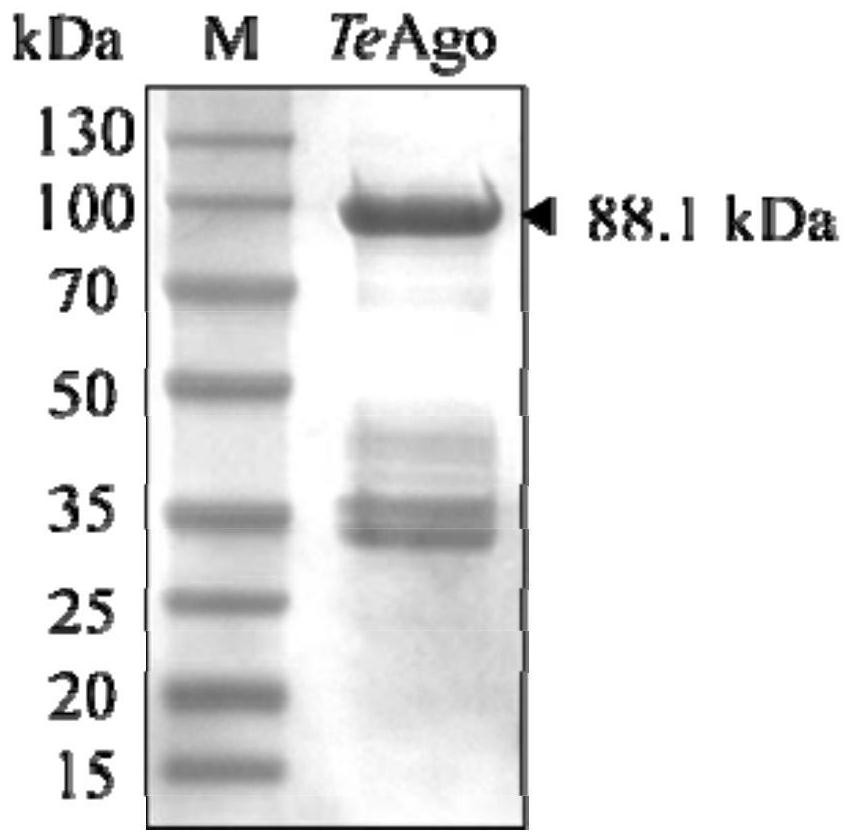
[0232] The above-mentioned TeAgo-pET28a prokaryotic expression plasmid was introduced into E.coli BL21(DE3) to obtain a TeAgo-pET28a / E.coli BL21(DE3) prokaryotic expression strain. The expression strain E.coliBL21(DE3) containing the recombinant plasmid TeAgo-pET28a was inoculated in LB medium containing 50 μg / mL kanamycin, and cultured on a shaker at 37°C and 220 rpm to OD 600 Between 0.6-0.8, add IPTG with a final concentration of 0.4-0.6mM, and continue culturing on a shaker at 200rpm at 18°C for 16-20h to induce the expression of TeAgo protein. Collect the cells by centrifugation, resuspend the cells in a resuspension buffer (containing 20mM Tris-HCl, pH around 8.0, 500mM NaCl), then crush the cells by high pressure, and centrifuge to obtain the supernatant. Using Ni-NTA column to affinity purify the protein, the eluate is concentrated by ultrafiltration, desalted and other steps to obtain the pu...
Embodiment 3
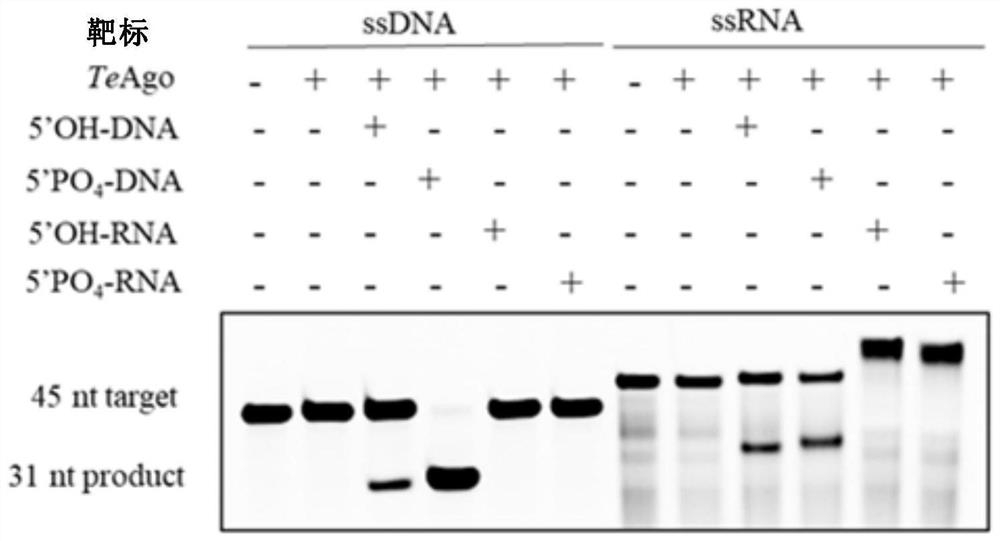
[0234] Example 3: TeAgo Shearing Activity Determination
[0235] Design fluorescently modified 45nt single-stranded DNA, RNA target nucleic acid and four complementary 16nt DNA and RNA guide strands, and send them to the company for synthesis.
[0236] DNA target nucleic acid sequence (SEQ ID NO:21):
[0237] 5'-FAM-CGCAGCATGTCAAGATCACAGATTTTGGGCTGGCCAAACTGCTGG-3'
[0238] RNA target nucleic acid sequence (SEQ ID NO:22):
[0239] 5’-FAM-CGCAGCAUGUCAAGAUCACAGAUUUUGGGCUGGCCAAACUGCUGG-3’
[0240] gDNA sequence (SEQ ID NO: 23):
[0241] 5'-HO / P-TAGTTTGGCCAGCCCA-3'
[0242] gRNA sequence (SEQ ID NO:24):
[0243] 5'-HO / P-UAGUUUGGCCAGCCCA-3'
[0244] Prepare the reaction buffer (containing 15mM Tris-HCl pH8.0, 250mM NaCl), and add MnCl with a final concentration of 0.5mM in the reaction buffer 2 , 400nM TeAgo, 2μM synthetic gDNA or gRNA and 0.8μM 5' fluorescently modified sequence complementary single-stranded DNA or RNA target nucleic acid, react at 95°C for 15min, af...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com