Method and device for detecting tumor neoantigen polypeptide
An antigen peptide, nascent technology, applied in bioinformatics, instrumentation, genomics, etc., can solve the problems of incomplete and inaccurate tumor neoantigen detection results, and achieve the effect of improving guiding significance and comprehensive sources.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
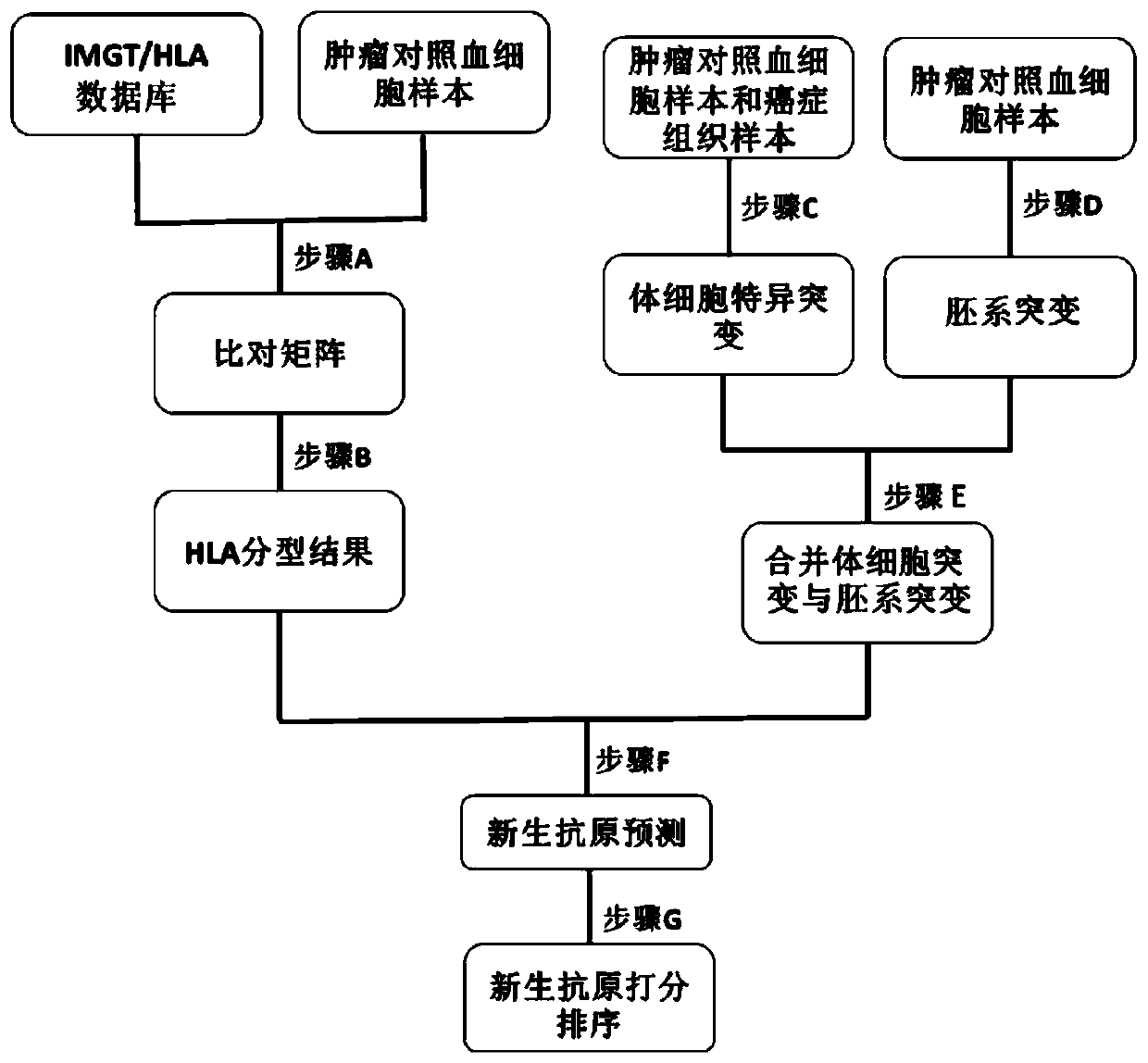
[0040] In a preferred embodiment of the present application, a method for detecting tumor neoantigen polypeptides is provided, figure 1 It is a flowchart of a method for detecting tumor neoantigen polypeptides according to an embodiment of the present invention. Such as figure 1 As shown, the method includes:
[0041] Step S101, obtaining somatic mutations and germline mutations of the tumor tissue;
[0042] Step S102, using the sequencing data of the tumor control blood cell sample to perform HLA typing to obtain the HLA typing result;
[0043] Step S103, using HLA typing results to predict neoantigen polypeptides for somatic mutations and germline mutations to obtain candidate neoantigen polypeptides;
[0044] Step S104, scoring and sorting the candidate neoantigen polypeptides, and the one with the highest score is the neoantigen polypeptide.
[0045] In the above method, the neoantigen polypeptide prediction is performed by obtaining the sum of mutations including soma...
Embodiment 2
[0068] Objective: To detect neoplastic antigens in tumor patients (sample ID 180504502TT1).
[0069] step:
[0070] 1. Use the software optitype (v2.1.0) and the sequencing fastq file of the tumor patient's blood cell sample to perform HLA typing, and obtain the typing results (see Table 1).
[0071] 2. Use the Mutect2 module of GATK (v4.0.5.1) to pair the tumor tissue and blood cells of tumor patients to detect somatic mutations, obtain the somatic mutation vcf file, and select mutations that pass the filtering criteria for subsequent use.
[0072] 3. Use the HaplotypeCaller module of GATK (v4.0.5.1) to detect germline mutations in the blood cells of tumor patients, obtain germline mutation vcf files, and select mutations that pass the filtering criteria for subsequent use.
[0073] 4. Merge the somatic mutation file with the Germline mutation file using the CombineVariants module of GATK (v4.0.5.1).
[0074] 5. Use vep (v94.5) to annotate the merged file to obtain the gene...
Embodiment 3
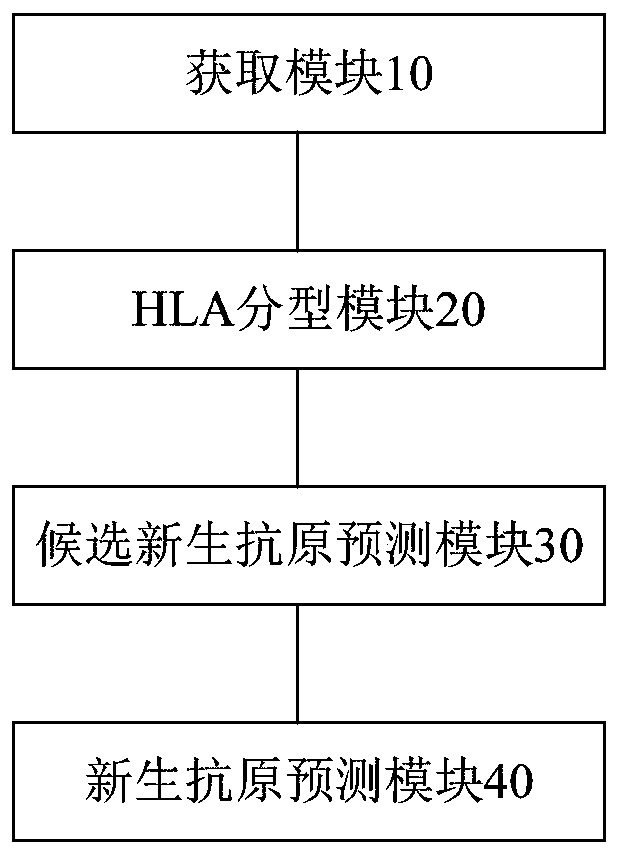
[0085] In this embodiment, a device for detecting tumor neoantigen polypeptides is also provided, such as image 3 As shown, the device includes: an acquisition module 10, an HLA typing module 20, a candidate neoantigen prediction module 30 and a neoantigen prediction module 40, and the acquisition module 10 is used to acquire somatic mutations and germline mutations of tumor tissues; The type module 20 is used to perform HLA typing using the sequencing data of tumor control blood cell samples to obtain HLA typing results; the candidate neoantigen prediction module 30 is used to use the HLA typing results to perform neoantigen classification for somatic mutations and germline mutations. Polypeptide prediction to obtain candidate neoantigen polypeptides; the neoantigen prediction module 40 is used to score and sort candidate neoantigen polypeptides, and record the highest score as neoantigen polypeptides.
[0086] The device predicts neoantigen polypeptides by obtaining the sum...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com