Method for constructing breast cancer neoadjuvant chemotherapy effect classification model
A classification model, a technique for breast cancer, used in medical reference, character and pattern recognition, healthcare informatics, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
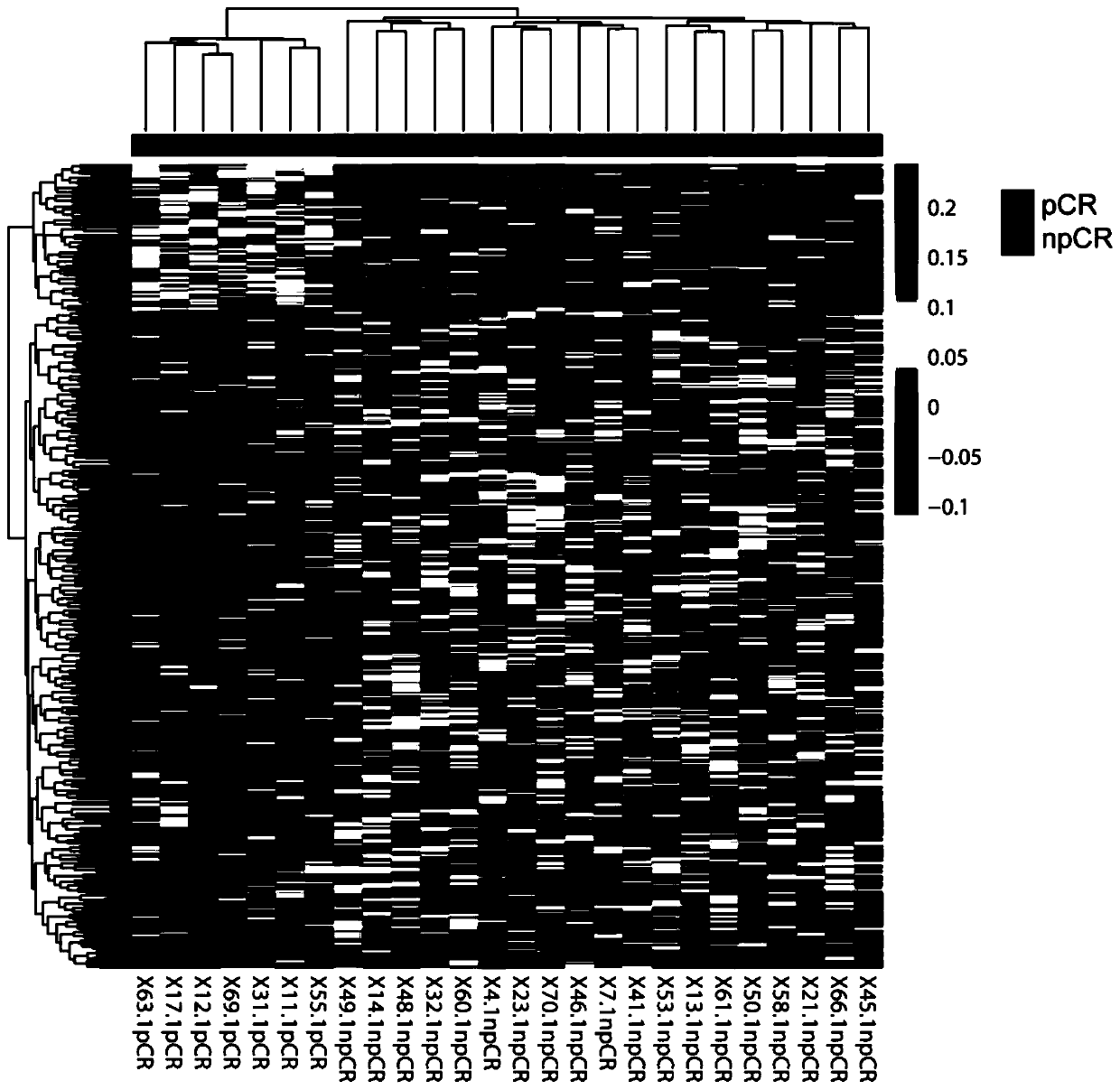
[0059] Example 1 Prediction of curative effect of the combination regimen of epirubicin, cyclophosphamide and paclitaxel in neoadjuvant chemotherapy for breast cancer
[0060] Step 1: Sample collection: collect peripheral blood from 26 breast cancer patients before neoadjuvant chemotherapy with epirubicin, cyclophosphamide, and paclitaxel, and evaluate the curative effect after 8 cycles of chemotherapy, according to RECIST1.0 Evaluation criteria, including 18 cases achieved PR effect, 8 cases achieved SD effect, 7 cases reached pCR according to whether pCR standard was met, and 19 cases did not reach pCR (npCR), as shown in Table 1.
[0061] Step 2: Plasma separation: The collected peripheral blood was centrifuged at 4°C and 1600g for 10 minutes, and the supernatant was centrifuged at 4°C and 16000g for 10 minutes for plasma separation. Be careful not to absorb white blood cells.
[0062] Step 3: Plasma cfDNA extraction: Plasma cfDNA was extracted using a plasma free DNA extra...
Embodiment 2
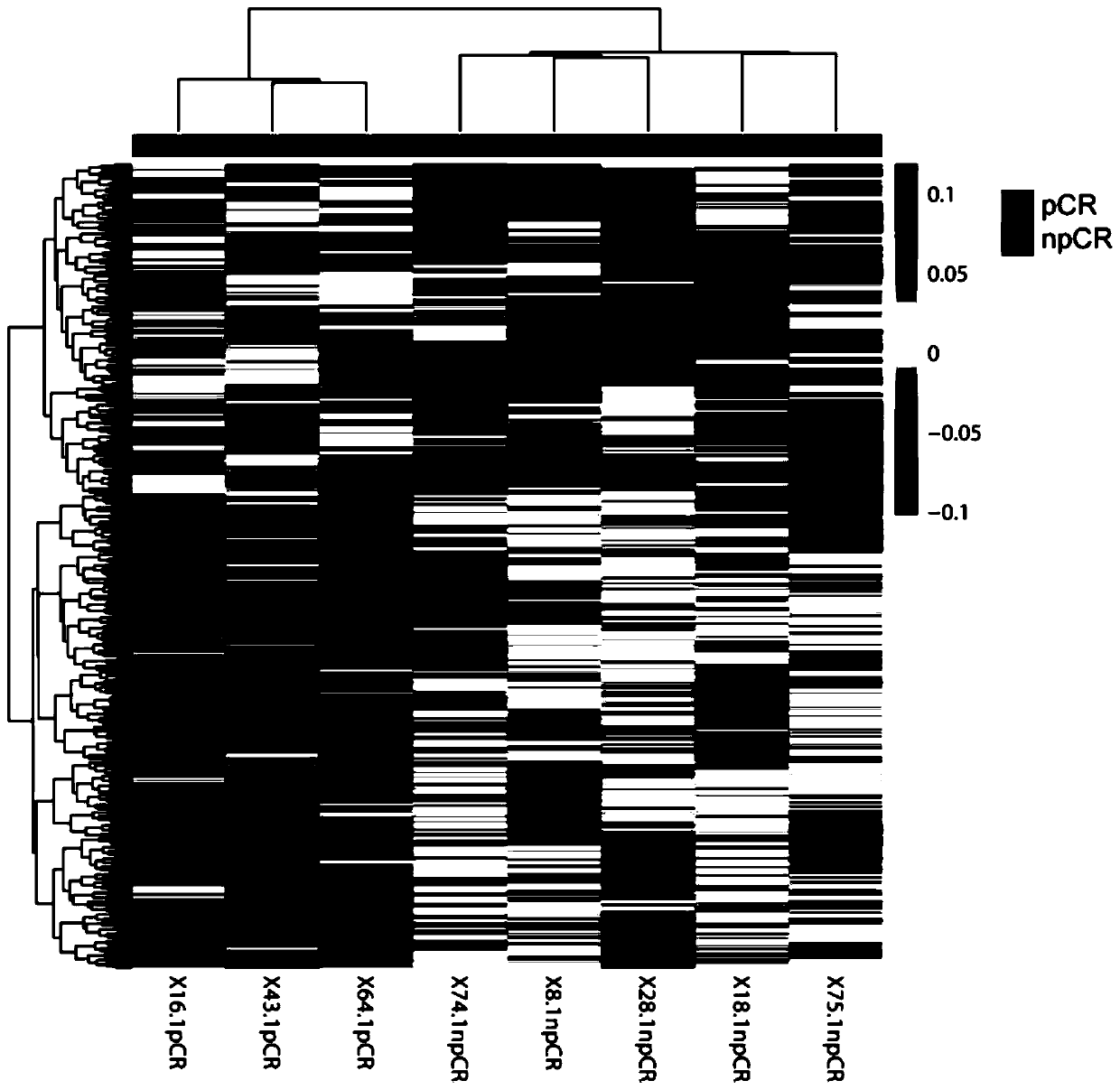
[0074] Example 2 Efficacy Prediction of Neoadjuvant Chemotherapy for Breast Cancer Epirubicin, Cyclophosphamide, Paclitaxel, and Carboplatin
[0075] Step 1: Collect peripheral blood from 8 breast cancer patients before neoadjuvant chemotherapy with epirubicin, cyclophosphamide, paclitaxel, and carboplatin, and evaluate the curative effect after 8 cycles of chemotherapy. Among them, 3 patients achieved pCR There were 5 patients who did not reach pCR (npCR), as shown in Table 1.
[0076] Steps 2-6 are the same as steps 2-6 in Embodiment 1, and the grouping method is only according to the grouping method of pCR and npCR.
[0077] The result is as image 3 As shown, based on the high-throughput detection of plasma cell-free DNA from 8 breast cancer patients who received a combined regimen of epirubicin, cyclophosphamide, paclitaxel, and carboplatin, and analyzed the coverage of TSSs, it was found that the pCR group was significantly different from the npCR group. A total of 555...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com