Srm/mrm assay for the tubulin beta-3 chain (TUBB3) protein
A protein, protein technology, applied in the determination/inspection of microorganisms, preparation of test samples, labels used in chemical analysis, etc., can solve the problem of not being considered cancer cells, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
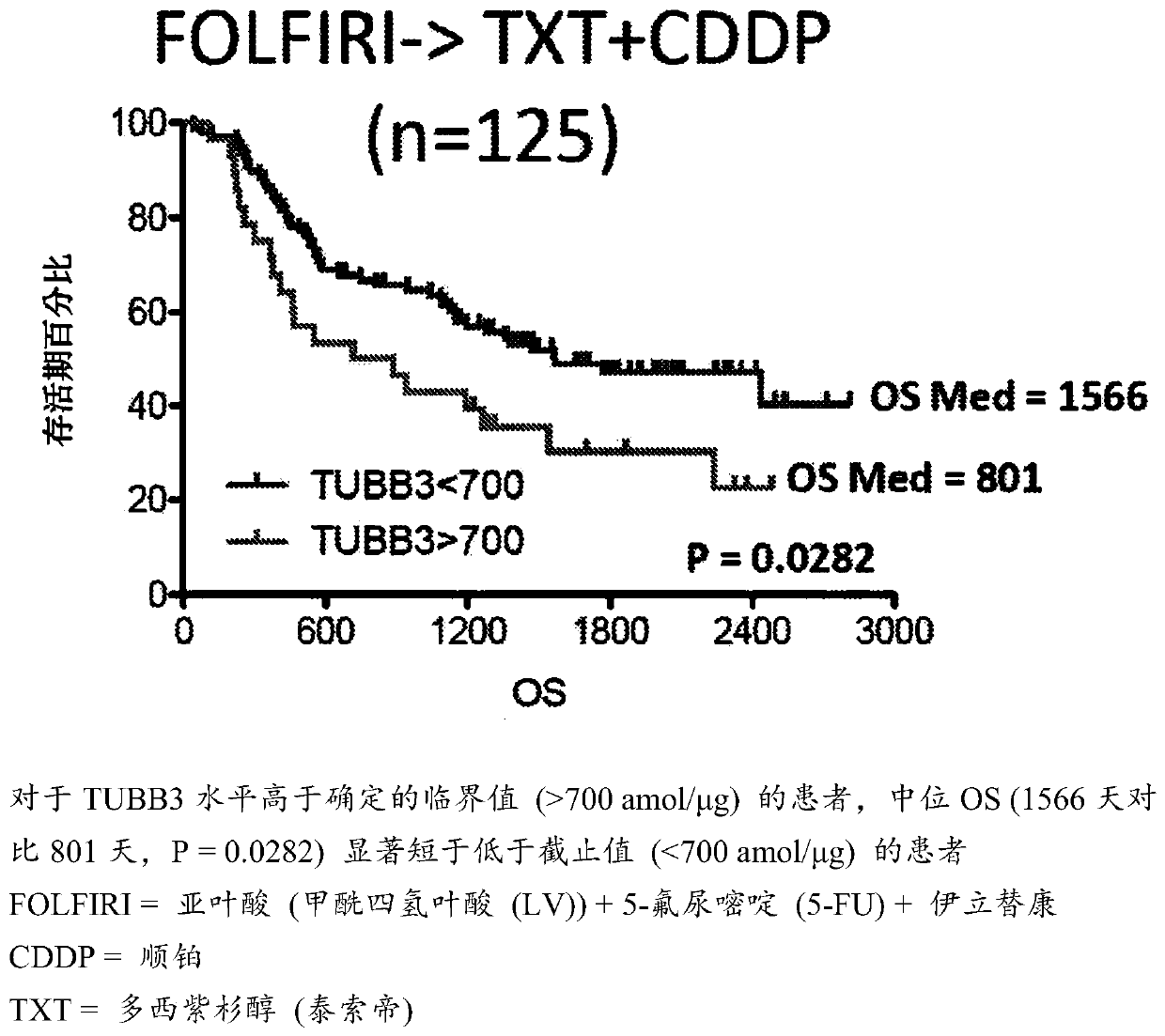
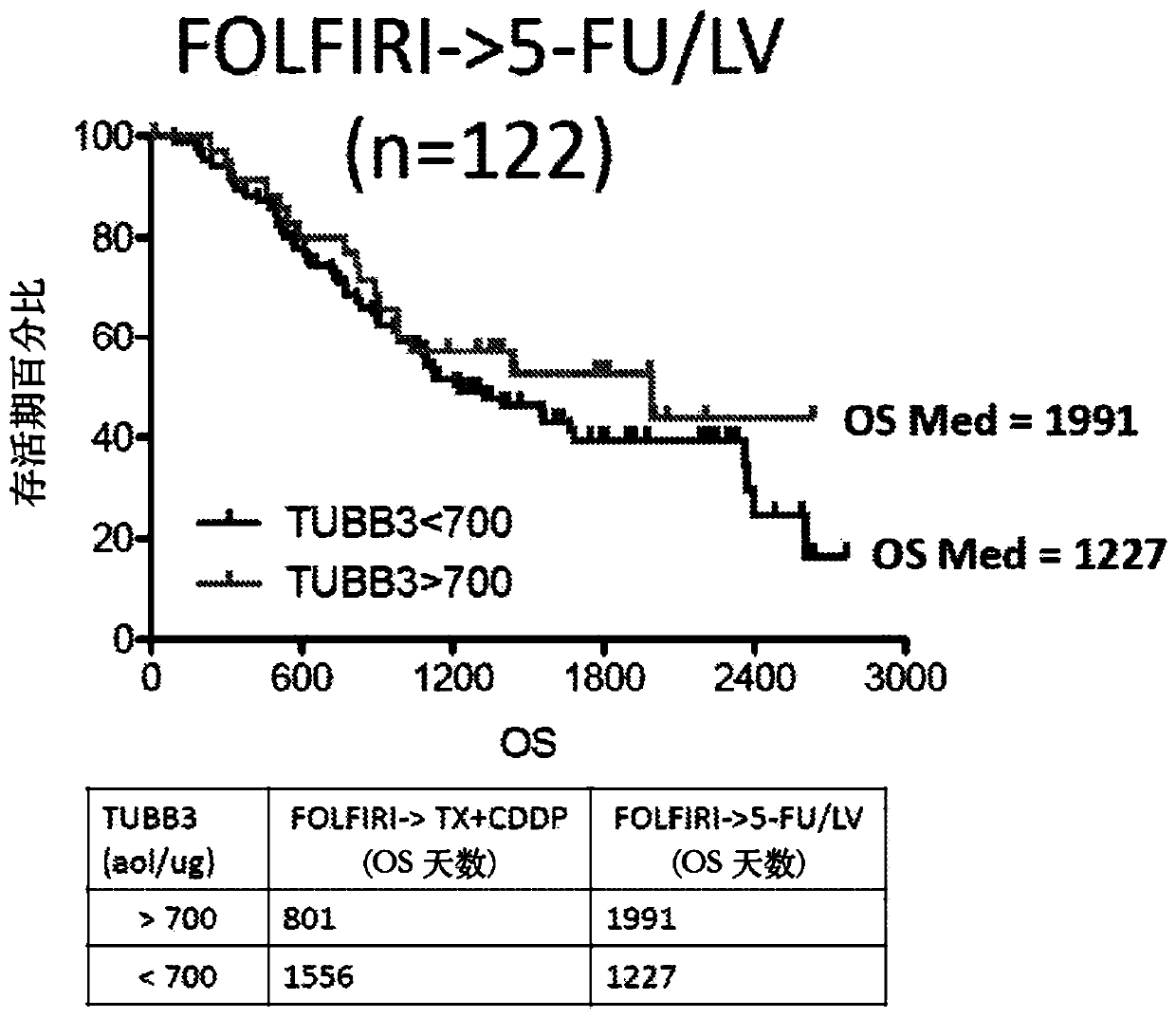
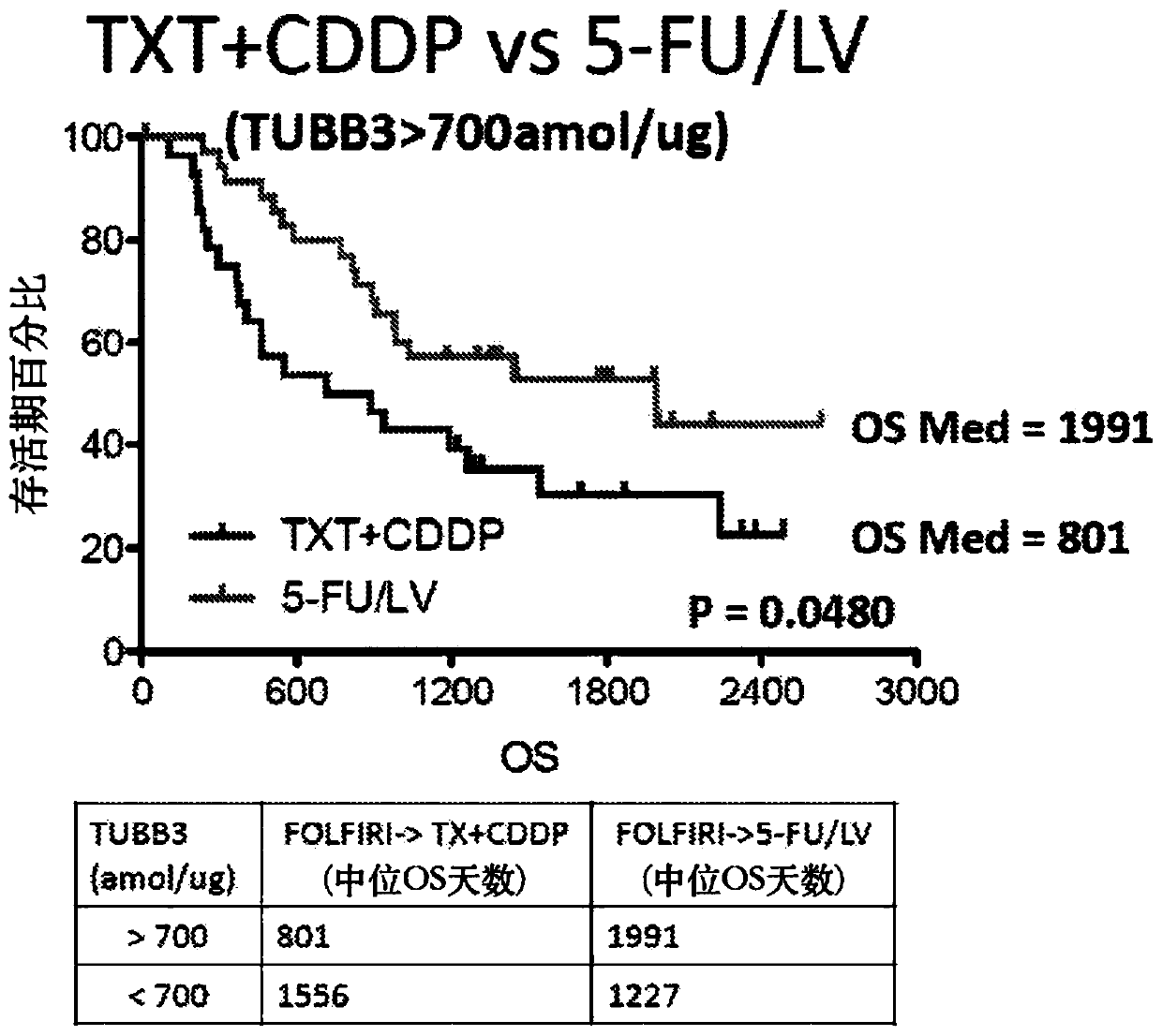
[0026] The assays described herein measure relative or absolute levels of specific unmodified peptides from the TUBB3 protein. Relative quantitative levels of TUBB3 protein can be determined by SRM / MRM methods, e.g., by comparing SRM / MRM signature peaks (e.g., signature peak areas or integrated fragmention intensities) of individual TUBB3 peptides in different samples . Alternatively, multiple SRM / MRM peak regions of multiple TUBB3 signature peptides, each with its own specific SRM / MRM signature, can be compared to determine the relative TUBB3 protein content in a biological sample , and compare it with the TUBB3 protein content in one or more additional or different biological samples. In this manner, the amount of one or more specific peptides from a TUBB3 protein is determined relative to the same one or more TUBB3 peptides across two or more biological samples under the same experimental conditions, and determined by The amount of this TUBB3 protein. Furthermore, compar...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com