Method for detecting target nucleic acid sequence amplified by multiplex amplification double signal
A nucleic acid sequence and nucleotide sequence technology, which is applied in the field of detection of target nucleic acid sequences amplified by multiplex amplification and double signal amplification, can solve the problem of time-consuming, limited types of fluorescent indicator molecules, and difficulty in realizing 5-plex detection. and other problems, to achieve the effect of excellent sensitivity
Active Publication Date: 2022-04-05
精确诊断有限公司
View PDF10 Cites 0 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Although the existing real-time PCR method has the advantage of being able to simultaneously perform amplification and detection in a homogeneous assay, the number of target nucleic acid sequences that can be detected simultaneously will also be limited because the types of fluorescent indicator molecules are limited. restricted
The existing thermal cycler (thermocycler) that can detect target nucleic acid sequences in real time can detect 5-plex at most at the same time, so the number of target nucleic acid sequences that can be detected at the same time will also be limited, and in order to test large-capacity It takes a lot of time to analyze the raw materials, and it needs to be equipped with an additional expensive real-time monitoring device
[0007] The most representative TaqMan probe method (US Patent No. 5,210,015) and the self-quenching fluorescent probe method (US Patent No. 5,723,591) in real-time PCR have false positive results due to non-specific binding of double-labeled probes Therefore, it is actually difficult to realize the detection of 5-plex and must have skilled technology and skills as support
Because the existing PCR method needs to perform amplification and detection at the same time, the real-time PCR device has limited high-speed processing (High Throughput) capabilities.
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
Embodiment 2
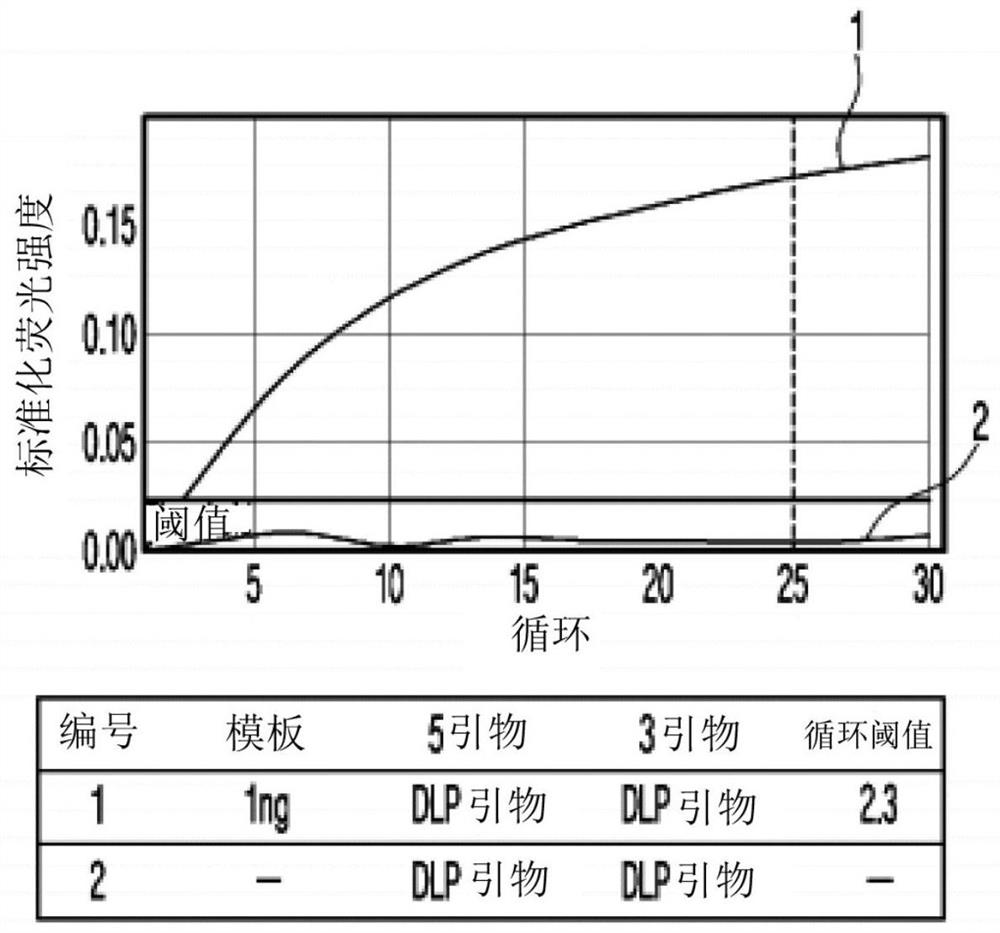
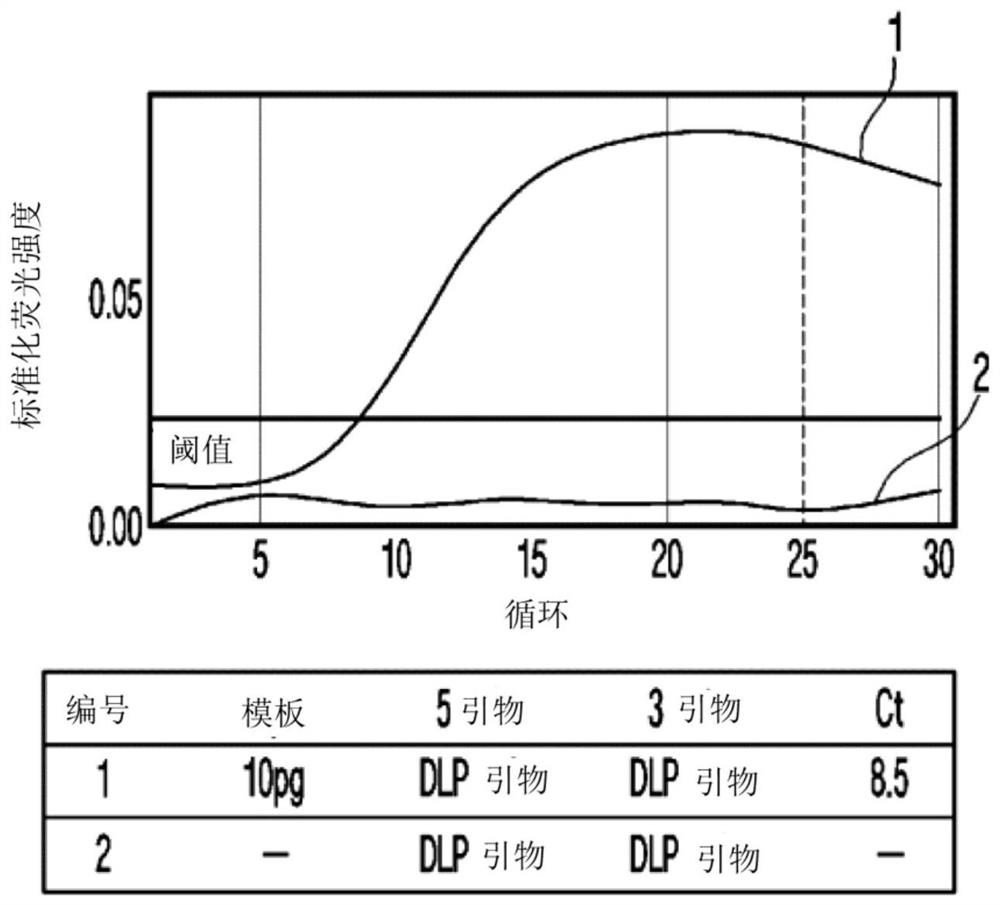
[0111] : PCR sensitivity applicable to the method of the present invention
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More PUM
 Login to View More
Login to View More Abstract
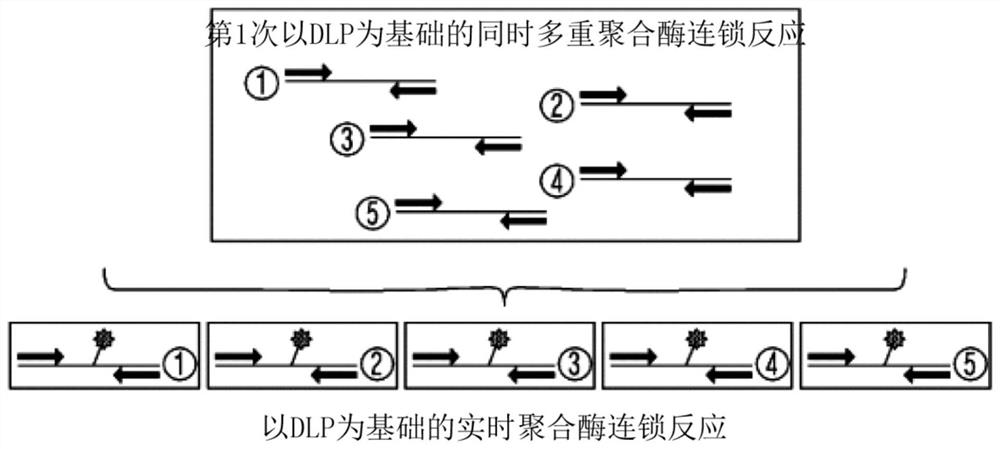
The present invention relates to a detection method (multiple amplification nested signal amplification; MANSA) of a target nucleic acid sequence amplified by multiple amplification double signals, in particular to a method for detecting a target nucleic acid sequence by using a dumbbell-shaped oligonucleotide for the first time A method for amplifying and detecting a target nucleic acid sequence through the amplification reaction and the second signal amplification reaction. The present invention can detect multiple target nucleic acid sequences in very limited samples without false positives.
Description
technical field [0001] The present invention relates to a detection method (multiple amplification nested signal amplification; MANSA) of a target nucleic acid sequence amplified by multiple amplification double signals, in particular to a method for detecting a target nucleic acid sequence by using a dumbbell-shaped oligonucleotide for the first time A method for amplifying and detecting a target nucleic acid sequence through the amplification reaction and the second signal amplification reaction. Background technique [0002] At present, the most common method used by researchers to obtain genetic samples is the polymerase chain reaction method using DNA polymerase. The oligonucleotides used in the polymerase chain reaction are designed to bind to opposite strands of the template DNA. Although the above method has the advantage of being able to accurately amplify only the desired fragment in the target gene by arbitrarily adjusting the length and base sequence of the olig...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More Patent Type & Authority Patents(China)
IPC IPC(8): C12Q1/6844
CPCC12Q1/68C12Q1/6848C12Q2525/119C12Q2525/301C12Q2527/107C12Q2537/143C12Q2549/119C12Q1/6876C12Q2525/101C12Q2525/204C12Q2600/16
Inventor 林成植金渊洙
Owner 精确诊断有限公司
Features
- Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com