Nanopore based dislocation sequencing method for direct sequencing of non-natural nucleic acid
A non-natural, nanopore technology, applied in the field of dislocation sequencing, can solve the problems of final sequence interference, errors, non-contiguous sequencing of non-natural nucleic acid strands, etc., and achieve low-cost results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1: Early Verification
[0035] Verification content mainly includes:
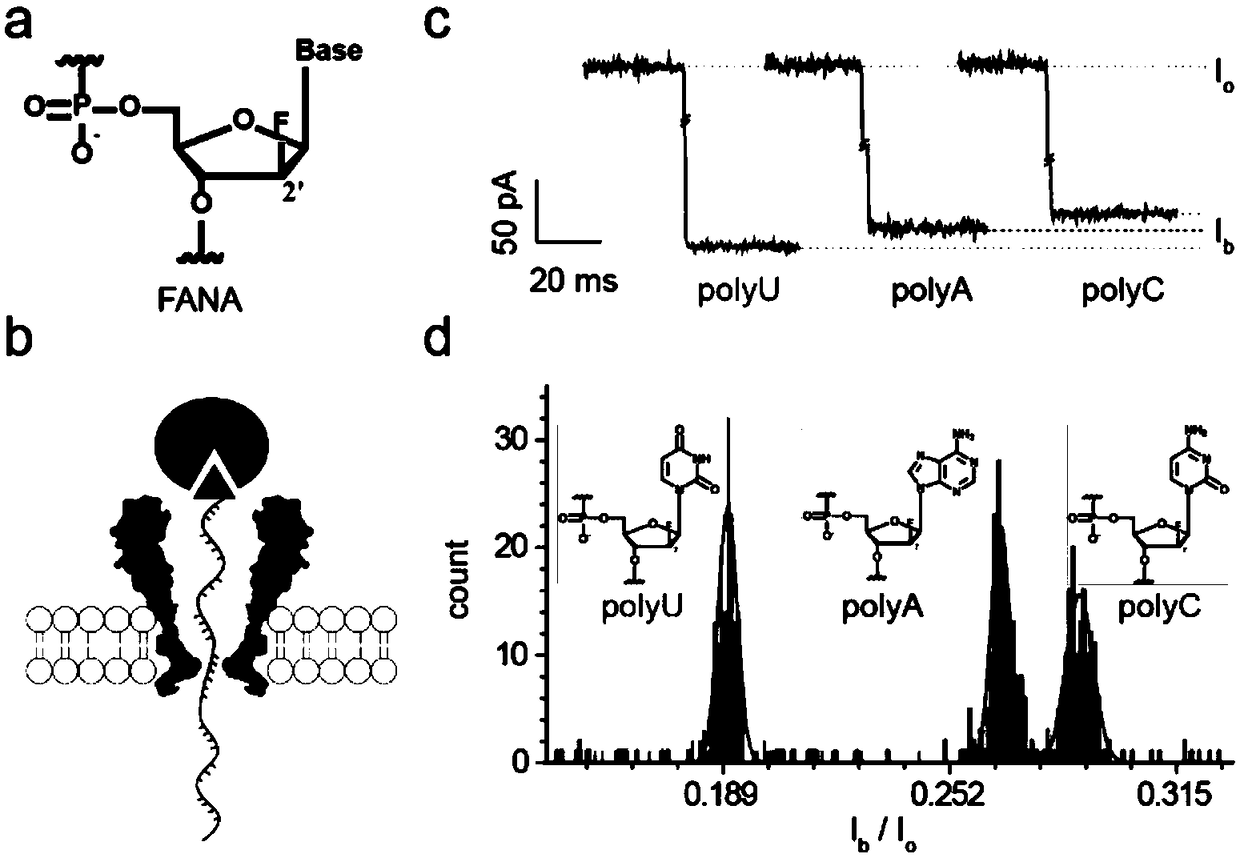
[0036] (1) Synthesize three single chains of A, C and U polymers of FANA labeled with biotin at the 3' end (Table 1, FANA polyA, FANA polyC, FANA polyU), which are used to statically simulate the pore blocking signal of a single FANA base , the result is attached image 3 shown.
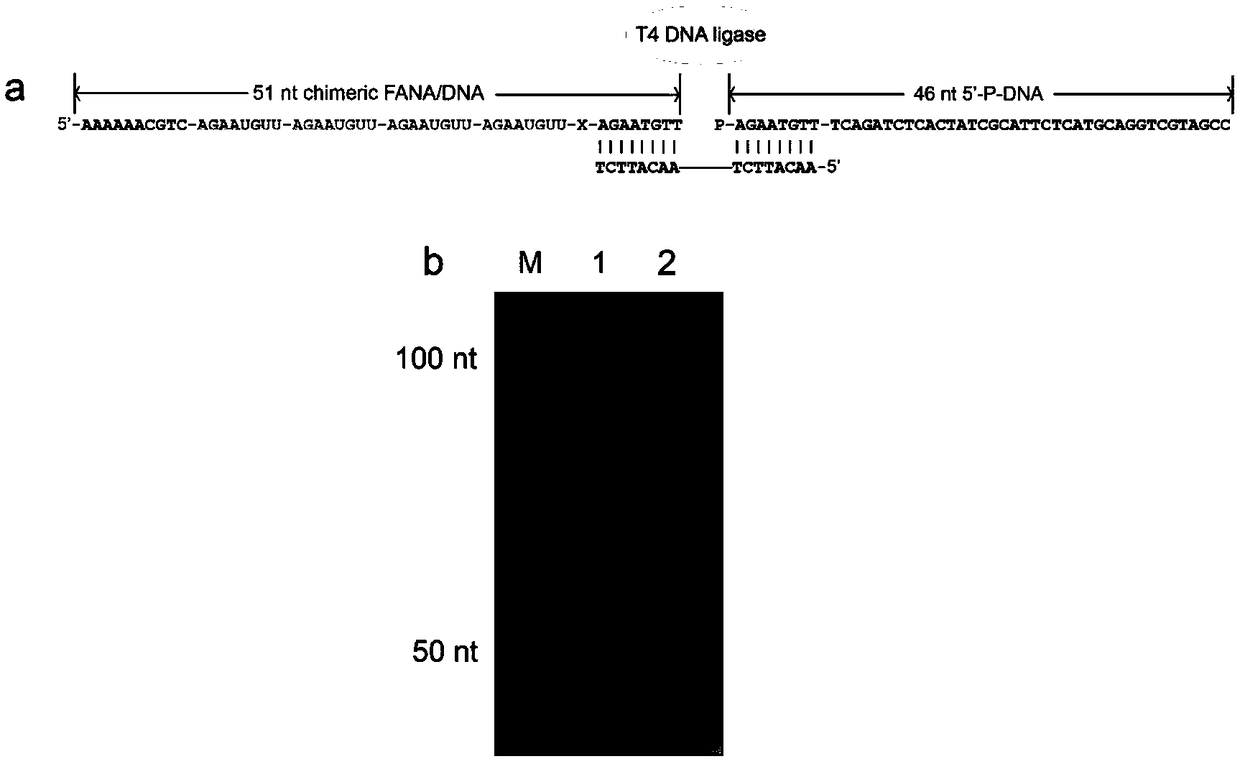
[0037] (2) Synthesize a FANA / DNA chimera (FANAx) with an abasic site monomer, and verify the feasibility of FANA sequencing, including in vitro reverse transcription experiments and sequencing signal reading. The results are shown in the attached Figure 4 shown.
[0038] The specific design process:
[0039] 1. Design the FANA single strand to be sequenced.
[0040] a. The A, C and U single strands of three FANAs labeled with biotin at the 3' end are used to statically simulate the pore blocking signal of a single FANA base. The results are shown in the attached image 3 shown.
[0041] b. Four FANA monomer co...
Embodiment 2
[0068] Embodiment 2: Sequencing example
[0069] Verification content mainly includes:
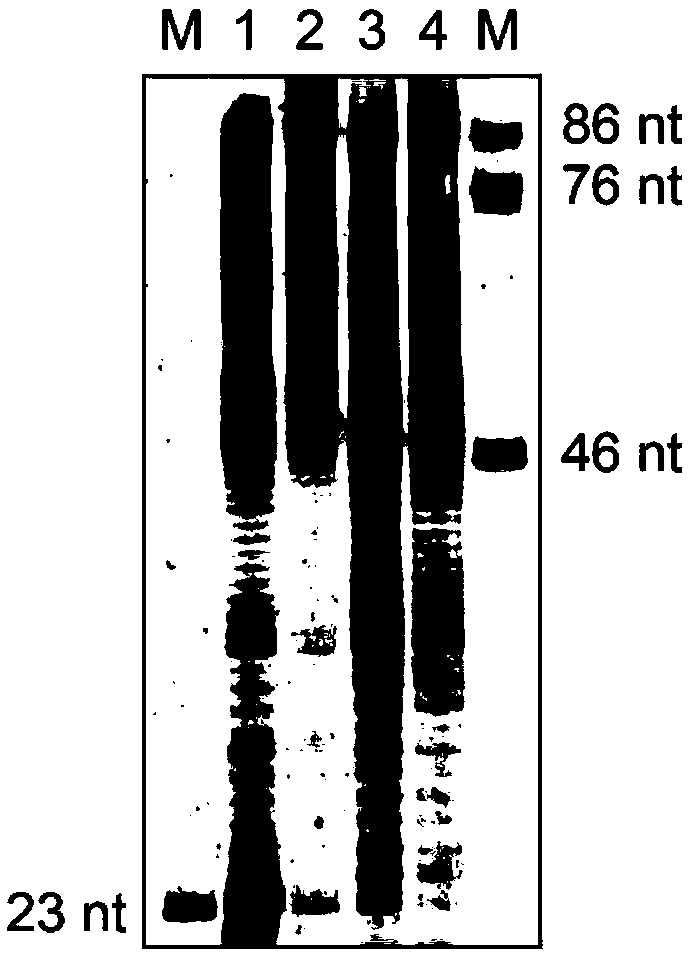
[0070] (1) Synthesize a random sequence FANA / DNA chimera (FANA30), compare the difference between DNA and FANA sequencing signals, including in vitro reverse transcription experiments and reading of sequencing signals, the results are as attached Figure 5 shown.
[0071] (2) Synthesize a modular FANA / DNA chimera (FANA42), study phi29DNAP as
[0072] The activity kinetics of FANA reverse transcriptase, including in vitro reverse transcription experiments and sequencing signal reading, the results are attached Figure 6 shown.
[0073] The specific design process:
[0074] 1. Design two DNA / FANA chimera sequences whose 3' end is a DNA monomer and the 5' end is a FANA monomer,
[0075] a. There are 30 randomly sequenced FANA monomers in the middle of the FANA30 chain.
[0076] b. In the FANA42 chain, the 5' end is a FANA monomer with 42 modular sequences.
[0077] 2. Using a DNA synth...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com