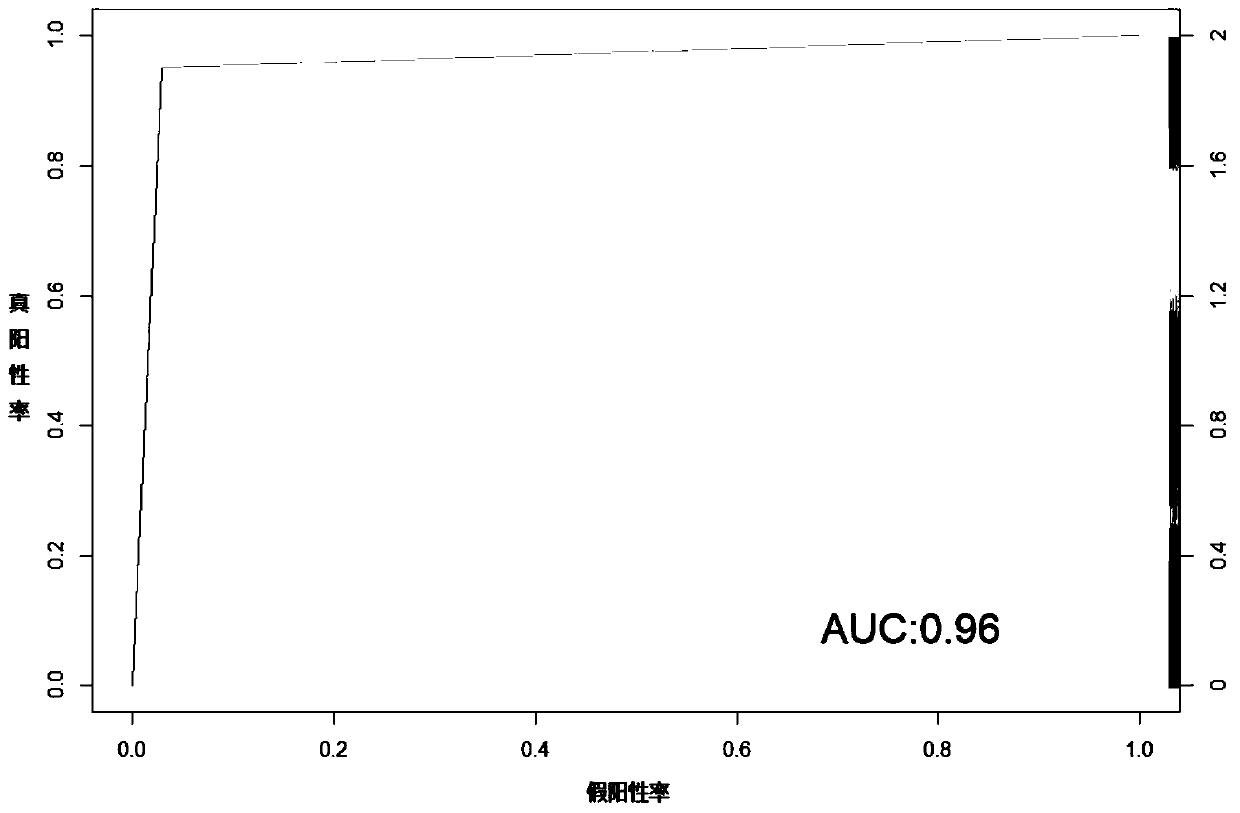
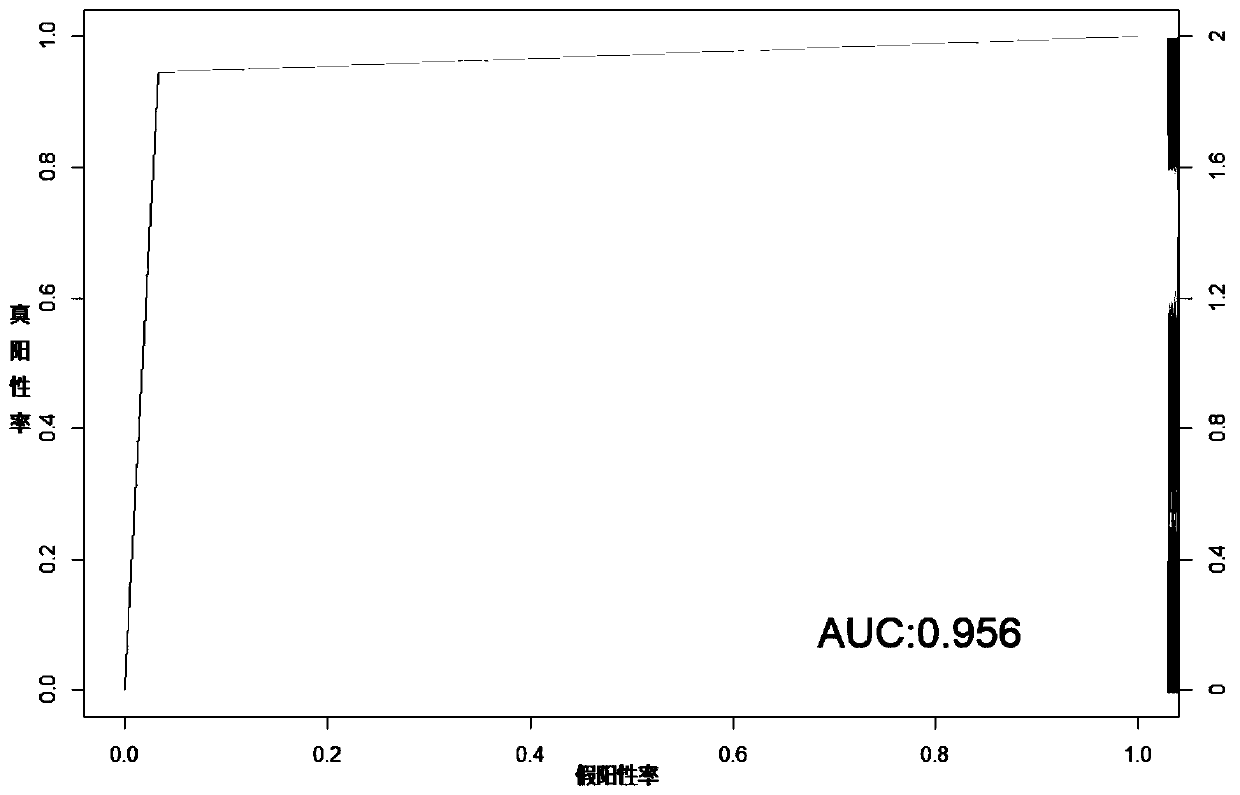
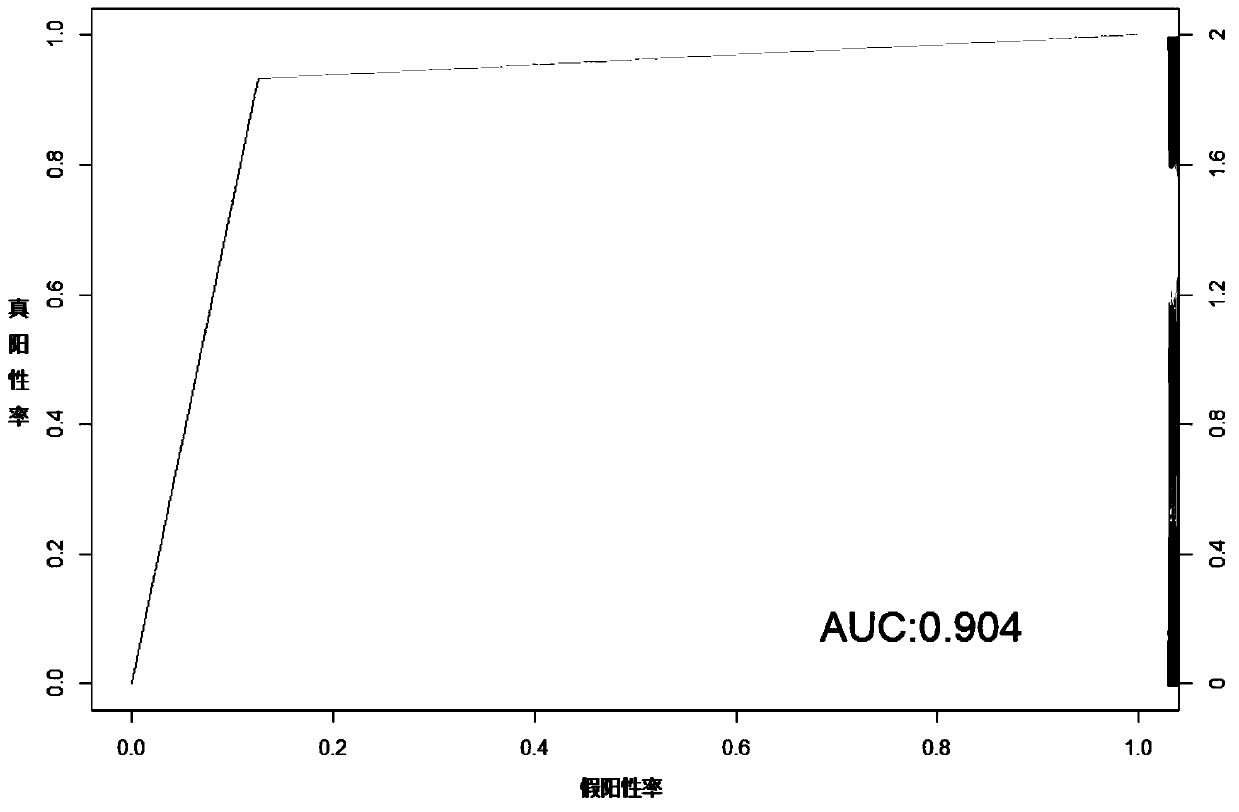
Molecular classification and application of multiple myeloma
A technology for patients with multiple myeloma and tumors, applied in the direction of instruments, biochemical equipment and methods, biostatistics, etc., can solve the problem that the plasma cell development process cannot be related, the cause of multiple myeloma has not been elucidated, and cannot be predicted Response to medication, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0067] Example 1, Screening of Molecular Diagnostic Markers for Multiple Myeloma and Implementation of Molecular Typing
[0068] Using the multiple myeloma expression data set GSE2658 provided by the NCBI GEO public database, 87 genes co-expressed with MCL1 were obtained through Pearson correlation analysis, and based on this, 46 genes with low expression of MCL1 were identified. -M genes enriched in multiple myeloma samples. In order to perform molecular typing more stably, 36 of the 133 genes with low classification efficiency were further screened out, and finally 97 classified genes with stable differential expression and relatively high abundance were retained.
[0069] The names of the 97 genes are as follows:
[0070] ACBD3, ADAR, ADSS, ALDH2, ANP32E, ANXA2, ATF3, ATP8B2, CACYBP, CAPN2, CCND1, CCT3, CDC42SE1, CERS2, CHSY3, CLIC1, CLMN, COPA, CSNK1G3, DAP3, DENND1B, ENSA, EPRS, EPSTI1, EVL, FAM13A, FAM49A, FLAD1, FRZB, GLRX2, HAX1, HDGF, HLA-A, HLA-B, HLA-C, HLA-F, HLA...
Embodiment 2
[0088] Example 2, Application of Bayesian Classifier for Multiple Myeloma in Predicting Patient Prognosis and Survival Rate
[0089] 1. Database GSE2658
[0090] According to the expression data of 97 classified genes of 551 cases of multiple myeloma patient samples (detection before treatment) in the GSE2658 database, the multiple myeloma Bayesian classifier obtained in Example 1 was used to classify the 551 cases of samples, and 249 samples were obtained. cases of MCL1-M-High subtype multiple myeloma and 302 cases of MCL1-M-Low subtype multiple myeloma.
[0091] 551 sample patients were followed up for 72 months after treatment, and survival analysis (K-M analysis and cox regression analysis) was carried out according to the follow-up results. The results are as follows: Figure 4 As shown, it can be seen that the two multiple myeloma subtypes, MCL1-M-High and MCL1-M-Low, have significantly different prognosis, and the overall survival rate of the MCL1-M-High subtype is hig...
Embodiment 3
[0100] Example 3. Molecular diagnostic markers and typing of multiple myeloma in predicting whether the patient to be tested can be treated with bortezomib
[0101] The GSE19784 multiple myeloma expression data set comes from a phase III drug clinical trial (HOVON-65 / GMMG-HD4), with the patient's drug treatment plan. In this trial, patients were randomly divided into two groups to receive two drug combinations of VAD (155 cases) and PAD (148 cases). The difference between the two is that the PAD program added bortezomib (trade name: Velcade). The gene expression data of the enrolled patients were collected before treatment.
[0102] The patients were stratified according to the above MCL1-M molecular classification, and then divided into MCL1-M-High (PAD 51 cases, VAD 56 cases) and MCL1-M-Low (PAD 104 cases, VAD 92 cases) subgroups. The survival analysis (K-M analysis and cox regression analysis) was carried out in groups according to the drug treatment regimen.
[0103] The...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com