A kit for detecting the mutation site of susceptibility gene for granular corneal dystrophy
A site and gene technology, applied in the field of kits for detection of mutation sites of susceptibility genes for granular corneal dystrophy, can solve the problems of long detection time, prone to false positives, and high cost, so as to reduce detection cost and shorten detection time time, the effect of rapid detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0136] Embodiment 1, be used to detect the preparation of the primer set of SNP site in TGFβI gene
[0137] The set of primers used to detect SNP sites in the TGFβI gene consists of primer pairs named Primer Pair A, Primer Pair B, and Primer Pair C, and extension primers named Extension Primer A1, Extension Primer A2, Extension Primer B, and Extension Primer C. Primer composition;
[0138] The primer pair A is composed of single-stranded DNAs named A-F and A-R respectively, and A-F and A-R are the single-stranded DNAs shown in sequence 1 and sequence 2 in the sequence listing, respectively;
[0139] Primer pair B is composed of single-stranded DNA named B-F and B-R respectively, and B-F and B-R are respectively the single-stranded DNA shown in sequence 3 and sequence 4 in the sequence listing;
[0140] Primer pair C is composed of single-stranded DNA named C-F and C-R respectively, and C-F and C-R are respectively the single-stranded DNA shown in sequence 5 and sequence 6 in ...
Embodiment 2
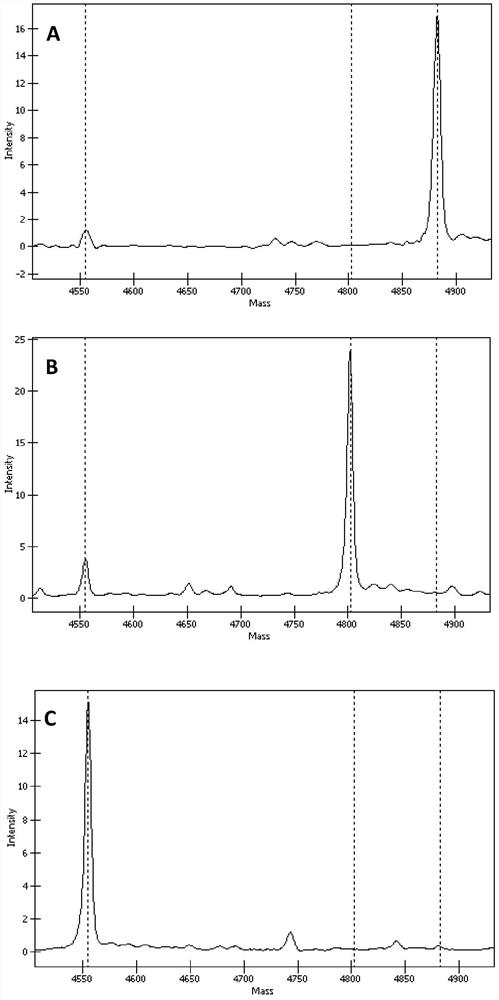
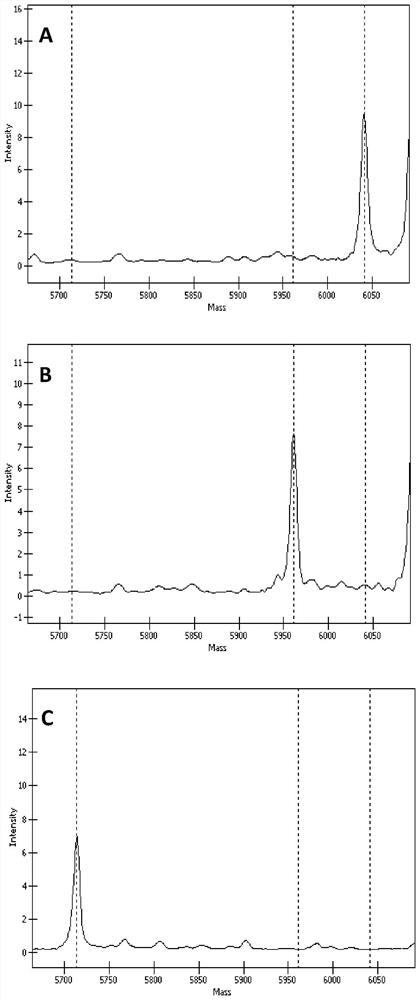
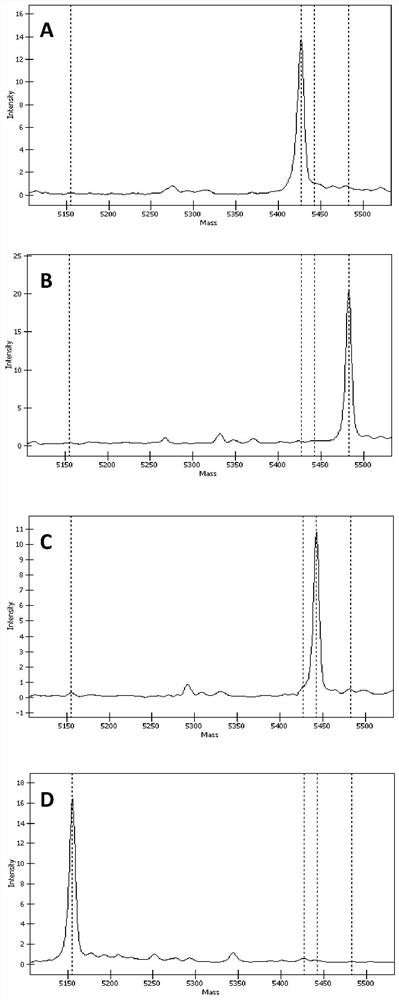
[0147] Example 2. Detection of TGFβI gene SNP sites——rs121909208, rs121909209, rs121909211 and rs121909215
[0148] Utilizing the set of primers in Example 1, using Agena Bioscience Inc.'s MassARRAY time-of-flight mass spectrometry biochip system and iPLEX Gold reagent (Agena Bioscience Inc.) to carry out genetic analysis on rs121909208, rs121909209, rs121909211 and rs121909215 of the blood samples of three people selected Typing detection, and preparation of these four sites for non-wild type (ie mutant) recombinant plasmids for verification. Among them, the nucleotides of the four SNP sites of the TGFβI gene of these three people were all detected and determined by the existing technology, and rs121909208, rs121909209, rs121909211 and rs121909215 were wild-type C, G, G and G, respectively. The experiment was repeated three times, and the specific steps are as follows:
[0149] 1) Genomic DNA extraction and sample preparation:
[0150] Extract genomic DNA from blood samples...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com