SNP locus related to lactation piglet diarrhea resistance or susceptibility, molecular marking detection and application
A piglet diarrhea, molecular marker technology, applied in the field of molecular genetics
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach 1
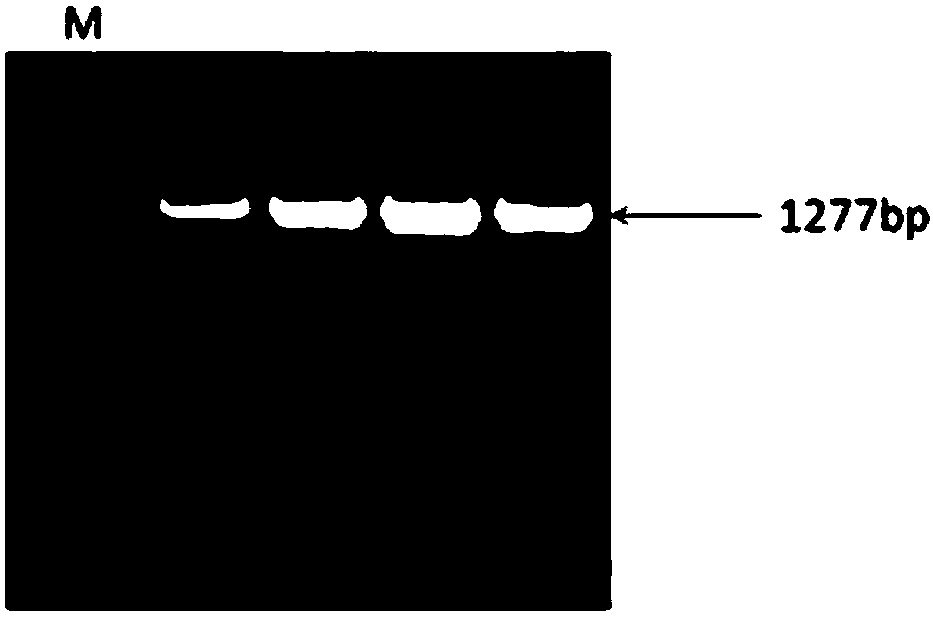
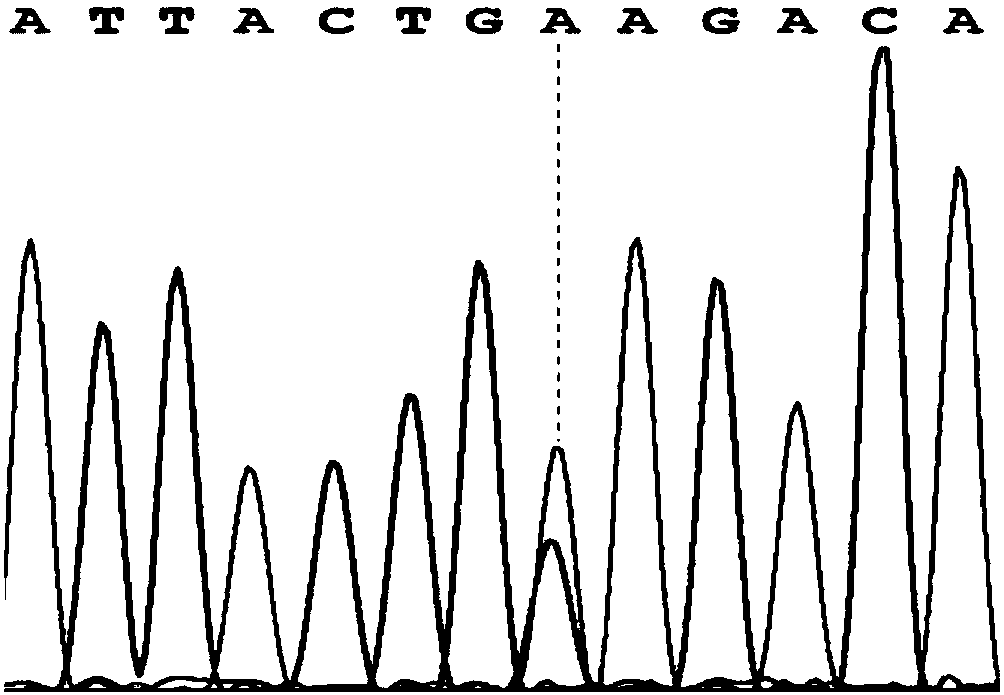
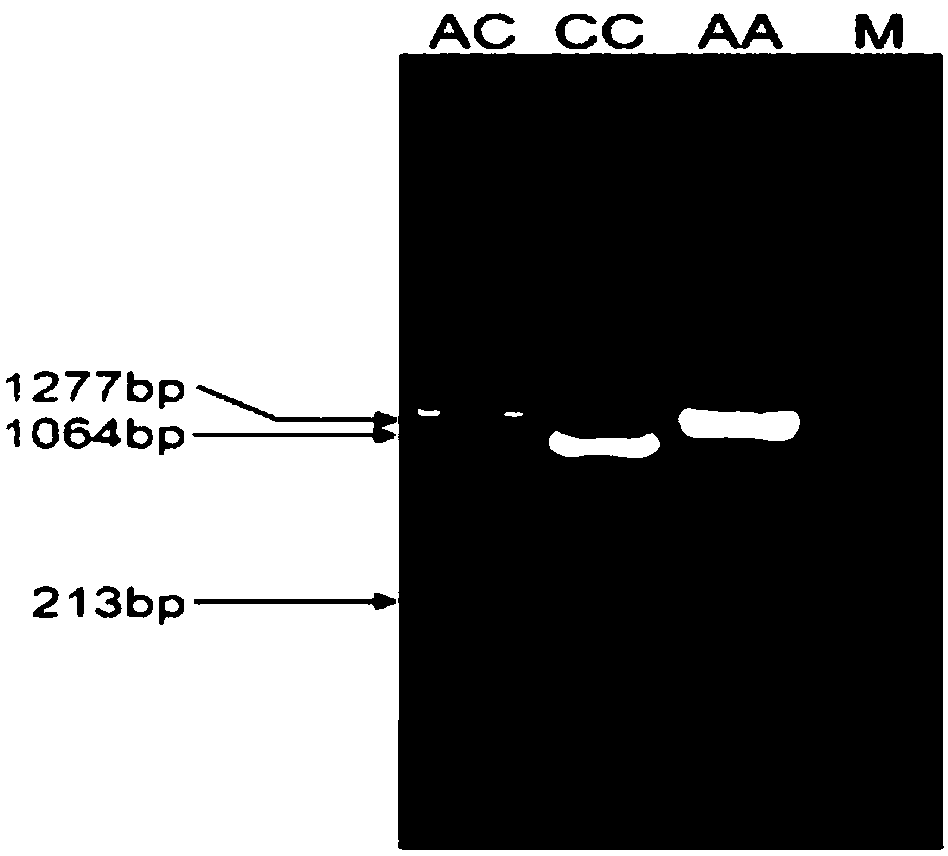
[0025] Specific embodiment one: A kind of SNP site related to suckling piglet diarrhea according to this embodiment, the SNP site is NC_010457.5:g.107288753C>A, located at 1063bp of the CTLA4 gene sequence, the CTLA4 gene sequence is as follows Shown in SEQ ID NO.1.
specific Embodiment approach 2
[0026] Embodiment 2: The molecular marker detection technology based on the SNP site described in Embodiment 1, the molecular marker detection technology is the PstI PCR-RFLP method.
specific Embodiment approach 3
[0027] Specific embodiment three: the sequence of the primer based on the PstI PCR-RFLP detection technology of the SNP site described in specific embodiment two is as follows:
[0028] C3F: 5'AGGTGCTTTCTCCTATTT 3'
[0029] C3R: 5'TTCTGTCTGTGGCATTAA 3'.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com