SNP nucleic acid mass spectrometric detection method for PLCE1 gene rs2274223 locus
A rs2274223-f, rs2274223-r technology, applied in the field of SNP nucleic acid mass spectrometry detection at the rs2274223 site of the PLCE1 gene, achieves the effects of strong reliability, guaranteed accuracy, and improved operability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0051] A method for detecting SNP nucleic acid mass spectrometry at the rs2274223 site of the PLCE1 gene, comprising the following steps:
[0052]S1: Perform the first round of PCR with the upstream primer rs2274223-F and downstream primer rs2274223-R to amplify the template DNA. The base sequences of the primers are as follows:
[0053] Upstream primer rs2274223-F: 5`-ACGTTGGATGCAAAGCCCATCCATCGAAAC-3`;
[0054] Downstream primer rs2274223-R:5`-ACGTTGGATGGATTCTCTGAGCAGTTACCG-3`;
[0055] The reaction system is as follows:
[0056]
[0057]
[0058] The reaction conditions are as follows:
[0059] (1) Pre-denaturation: 95°C for 2 minutes;
[0060] (2) Amplification: 95°C for 30s, 56°C for 30s, 72°C for 60s, a total of 45 cycles;
[0061] (3) Extension: 5min at 72°C, ∞ at 4°C;
[0062] S2: Dephosphorylate the amplified product obtained in step S1 to obtain a dephosphorylated fragment, and the SAP mixed liquid system used in the dephosphorylation operation is as follow...
experiment example 1
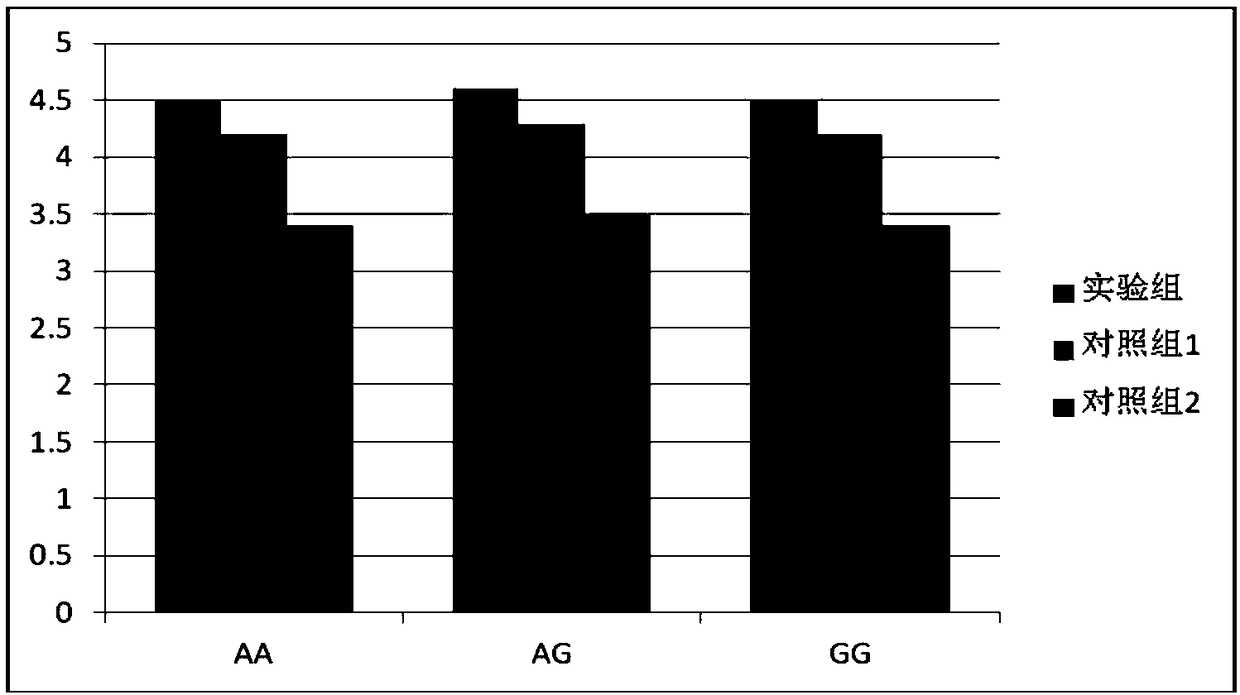
[0089] The method provided by the present invention is used as the experimental group, the method disclosed in Patent Publication No. 105238853A is used as the control group 1, and the conventional Taqman method is used as the control group 2, and the rs2274223 locus standard of the PLCE1 gene of 15 ng (including 5 ng GA hybrid combined, 5ng GG homozygous and 5ng AA homozygous) were detected, and the typing and quantitative results of each group of experiments were compared.
[0090] Such as figure 1 As shown, the typing results of the experimental group and the control group 1 were better than those of the control group 2, and the quantitative results of the experimental group were higher than those of the control group 1 and closer to the standard content. It shows that the SNP detection of the rs2274223 site of the PLCE1 gene by the method provided by the present invention is more accurate and more effective than the existing method, and it also shows that the denaturation-...
experiment example 2
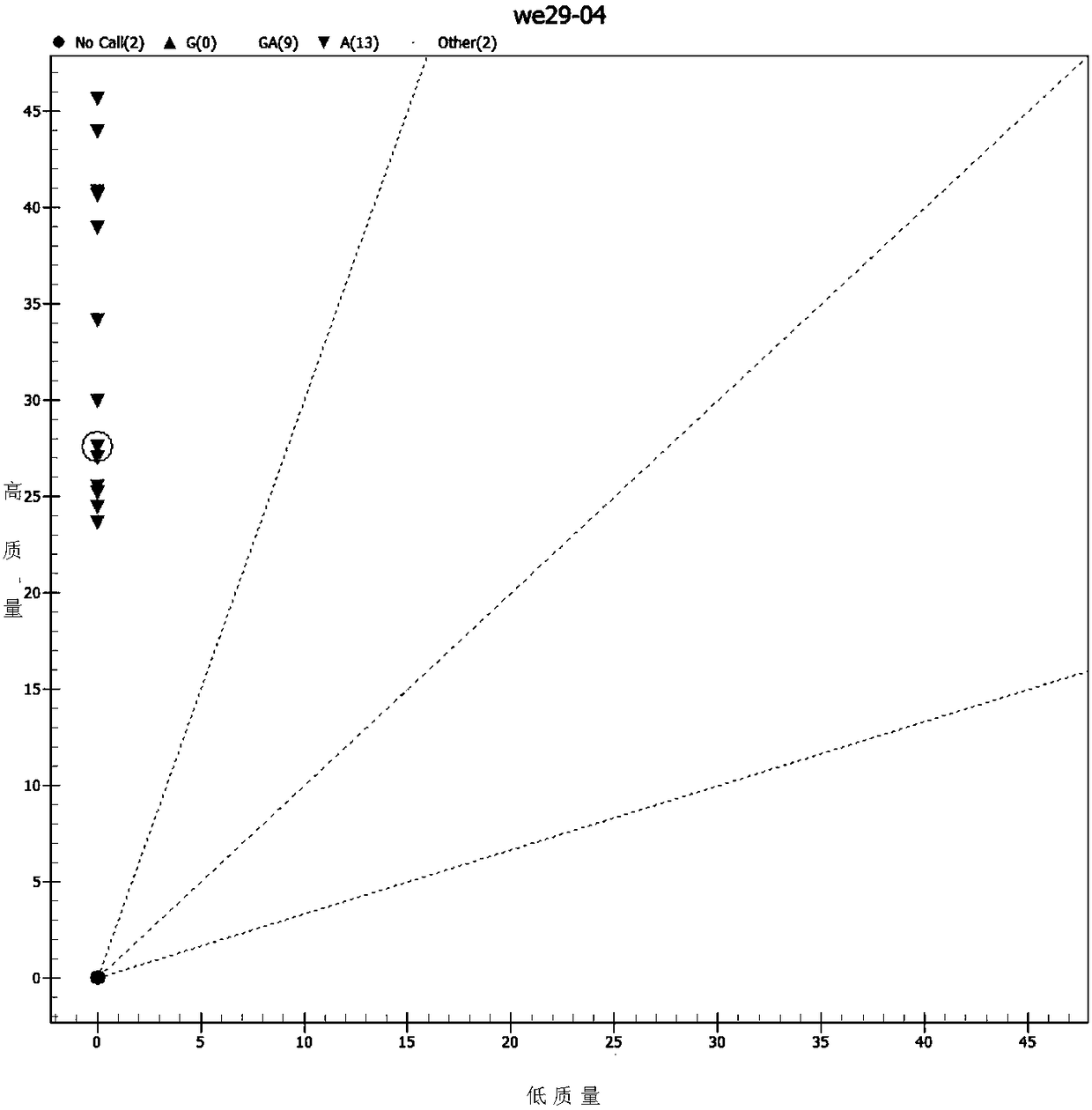
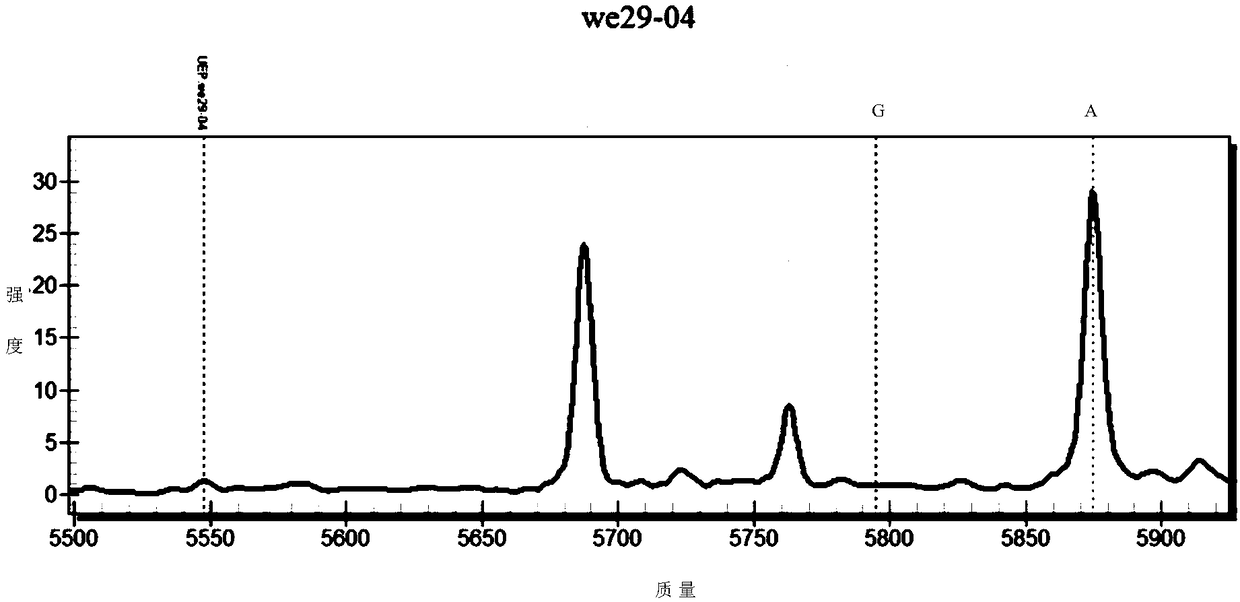
[0092] Randomly select 26 healthy adult volunteers (18-60 years old) to collect blood samples and extract the whole blood genome, and perform SNP detection at rs2274223 on 26 samples according to the method provided in the examples, and analyze the Mass spectrometry results were analyzed.
[0093] Experimental results such as Figure 2-4 As shown, a total of 22 samples obtained analyzable results, of which 9 samples were heterozygous for GA and 13 samples were homozygous for AA. It shows that there are more AA homozygotes at this SNP site, and G-A heterozygotes are also detected. figure 2 The sample points of different genotypes are separated from each other and have no intersection with each other. Figure 3-4 The peak types and separation conditions of different types in the test are good, indicating that the primers provided by the present invention have good specificity, high sensitivity, and accurate typing results.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com