A hybridization capture kit and method for detection of 16 gene hotspots in individualized medicine for tumors
A technology for hybridization capture and detection reagents, which is applied in biochemical equipment and methods, and microbial determination/inspection, etc. It can solve the problems of complex operation process capture technology, large fragment deletion of gene rearrangement cannot be detected, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0085] A hybridization capture kit and its method for detecting 16 gene hotspots in personalized medicine for tumors according to the present invention will be described in detail below in conjunction with the accompanying drawings and specific examples.
[0086] Hybridization capture kit for detection of 16 gene hotspots for individualized tumor medicine, including hybridization reagents, PCR amplification reagents and enrichment detection reagents.
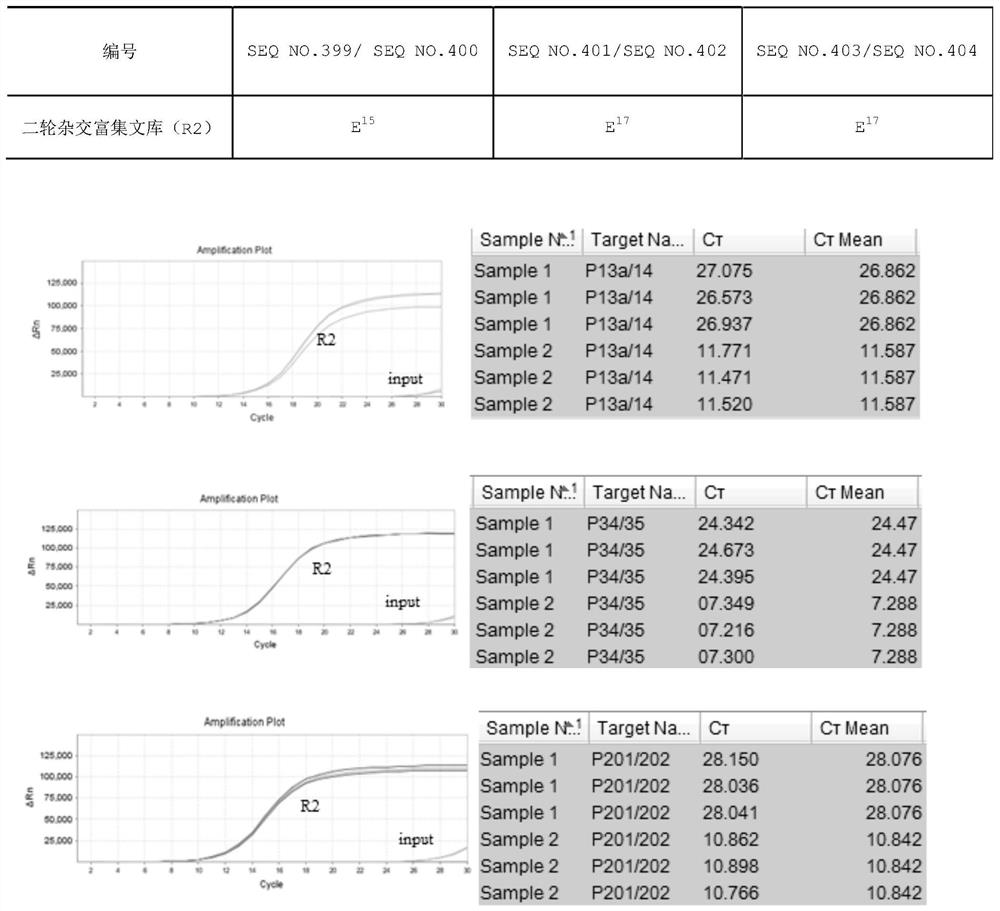
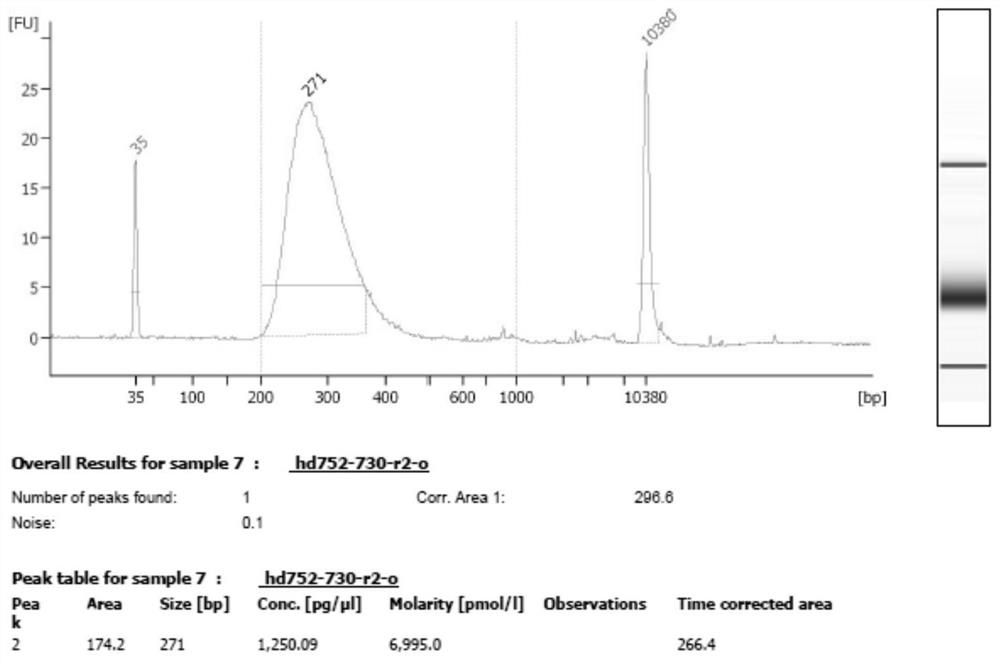
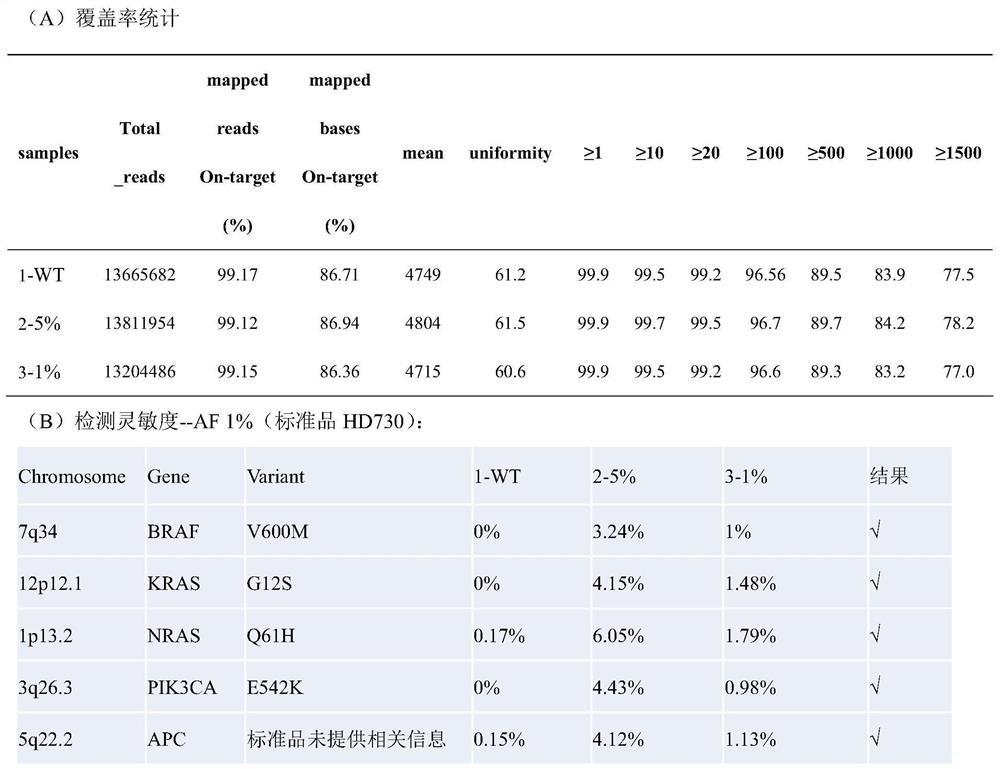
[0087] 1) Hybridization capture: the hybridization reagent includes a probe mixture (SEQ NO.1-SEQ NO.396), and the molar ratio of each probe is 1:1. For detection, the amount of the above-mentioned probe mixture with a concentration of 10 nM each was 1 μl. The hybridization capture kit for detection of 16 gene hotspots for individualized tumor medicine also includes reagents for the hybridization system: Block3.1 / 3.2 (SEQ NO.405 and SEQ NO.406), Human Cot-1 PerfectHyb TM Plus hybridization buffer. The molar ratio of Block3....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com