Target polynucleotide editing method and application thereof
A technology for targeting polynucleotides and oligonucleotides, which is applied in genome editing methods and its applications, and in the field of targeted cutting of genomes, which can solve the problems of restricting the application of artificial endonucleases and the limitation of RGEN targets
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] Example 1 Construction and expression of recombinant structure recognition endonuclease
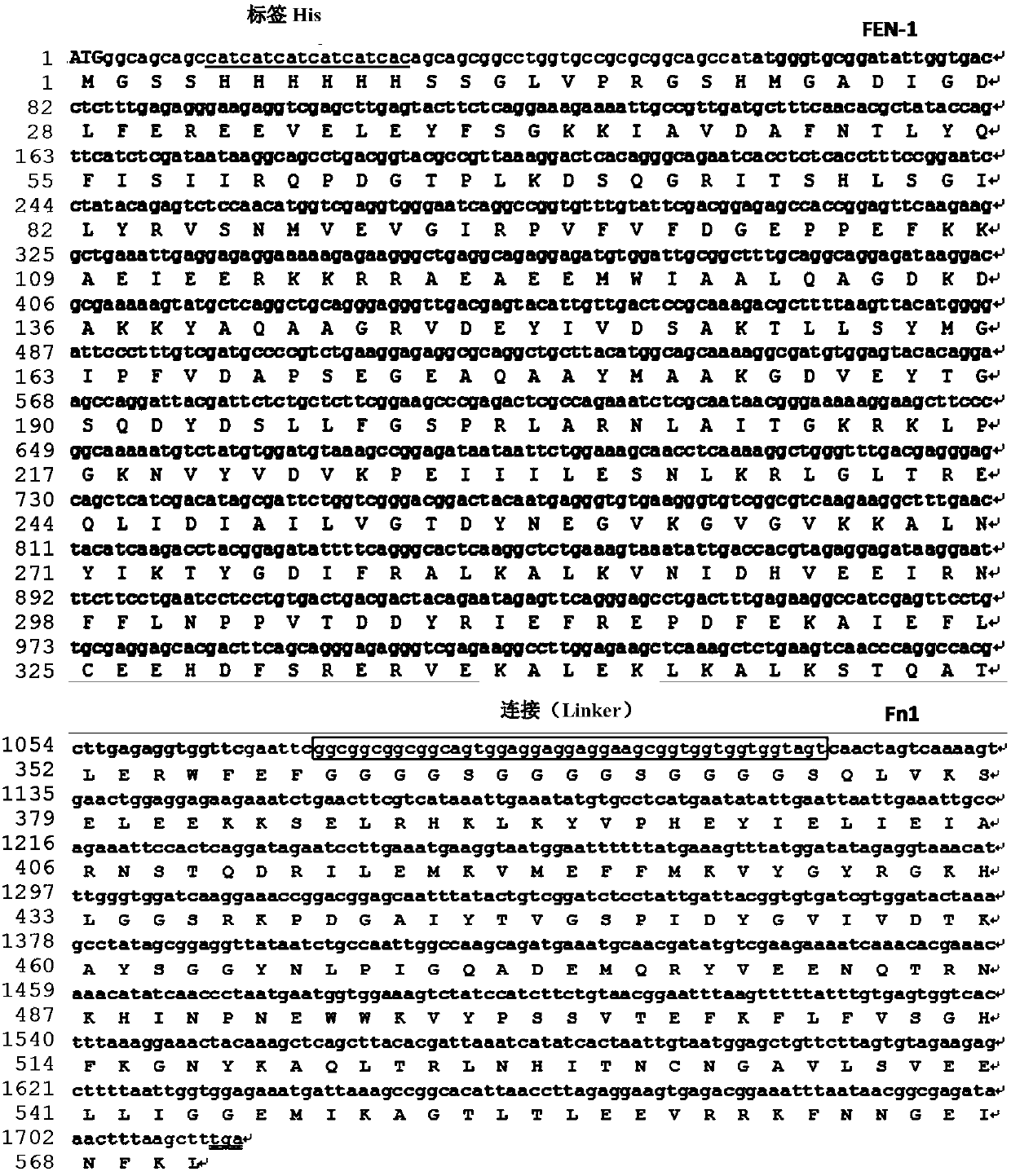
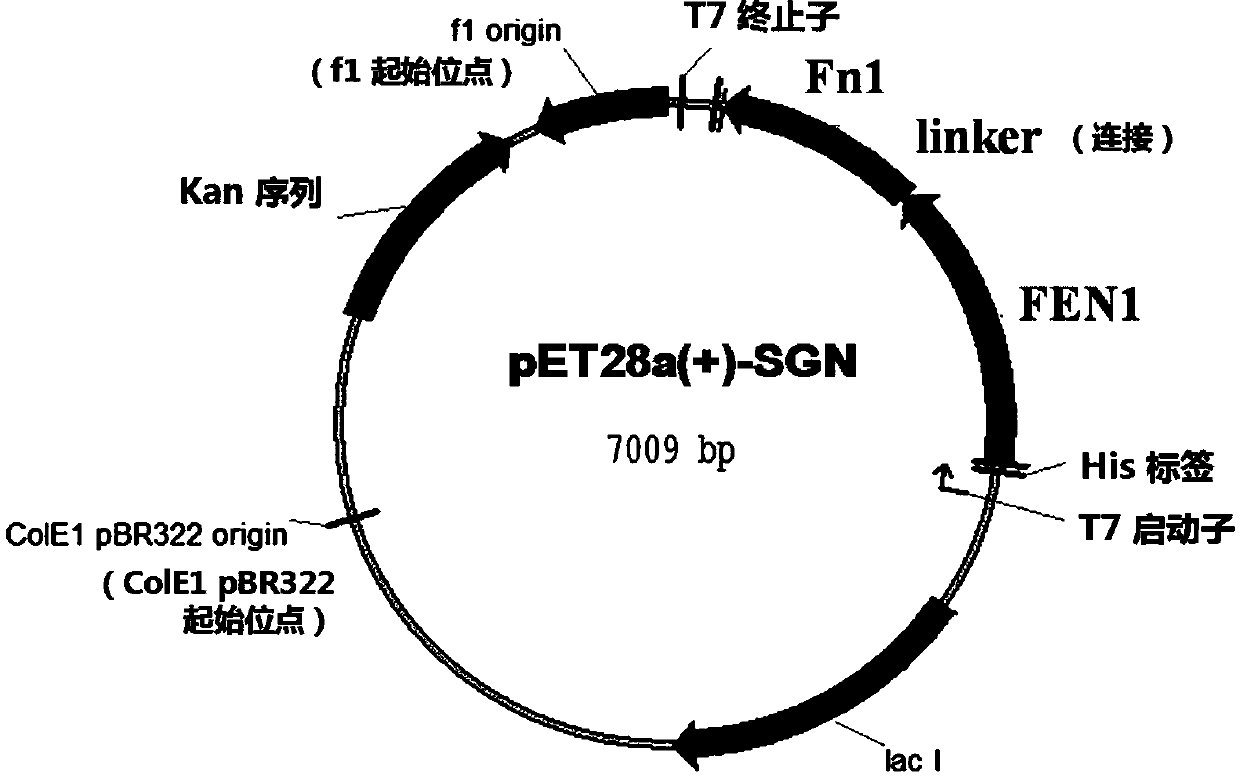
[0059] In the present invention, a recombinant structure recognition endonuclease (Structure-guided endonuclease, SGN) is constructed, which is composed of FEN-1 that recognizes the 3'-Flap structure and the Fok I (Fn1) cleavage domain that cuts the DNA chain . Wherein, the sequence is encoded by the following gene: Fok I (196 amino acid residues) at the C-terminal, glycine-serine repeat sequence at the middle link, and FEN-1 enzyme. This sequence was inserted into the prokaryotic expression vector pET28a(+) to form pET28a(+)-SGN. The coding sequence and amino acid sequence of SGN are shown in figure 1 , Figure 3-4 The plasmid map of pET28a(+)-SGN is shown in figure 2 shown in . In the pET28a(+)-SGN construct, the SGN gene is located downstream of the T7 promoter.
[0060] CaCl for pET28a(+)-SGN 2 Heat shock method for transformation into the host bacterial strain Arctic E...
Embodiment 2
[0064] Example 2 SGN cuts single-stranded DNA in vitro
[0065] To test whether the engineered SGN can cleave DNA strands, we incubated 1 ng SGN, 10 pmol substrate single-stranded DNA (ssDNA) (S-1), and 10 pmol gDNA-1 in a 10-μL reaction (all ssDNA and DNA The sequence of the oligonucleotide gDNA is shown in Table 1), and the 10-μL reaction system also included MOPS (10mM), 0.05% Tween-20, 0.01% nonidet P-40 and MgCl 2 (7.5mM). Among them, the 5' end of S-1 is labeled with a fluorescent Cy5 group. And before SGN was added, the mixture was first incubated at 95°C for 5 minutes and at 55°C for 10 minutes. Then add SGN and react at 37°C for 2 hours.
[0066] The gDNA-1 and S-1 form a 3'-Flap structure. SGN was added to the mixture for reaction, and then the mixture was separated by denaturing-PAGE and imaged by fluorescence. The specific steps are that the product obtained by the reaction is analyzed by PAGE under denaturing conditions. Loading buffer contained 90% formamid...
Embodiment 3
[0072] Example 3 SGN cleavage activity is not dependent on the target sequence
[0073] In order to prove whether SGN has DNA sequence preference, ssDNAs with different sequences (S-2, S-3, see Table 1 for sequence) were used as the substrate of SGN for reaction. The 10-μL reaction system and conditions are the same as those described above with S-1 as the substrate.
[0074] The results showed that, when guided by gDNA-2 or gDNA-3, SGN could cut S-2 or S-3 respectively (see Image 6 b and Image 6 Lane 5 of c). while only containing S plus SGN ( Image 6 b and Image 6 Lane 1 of c), S plus gDNA ( Image 6 b and Image 6 Lane 2 of c), S plus Fok I and gDNA ( Image 6 b and Image 6 Lane 3 of c), or S plus FEN-1 and gDNA ( Image 6 b and Image 6 No cleavage occurred in the reaction of lane 4) of c. This result indicates that SGN cleavage activity is independent of the target sequence but recognizes the 3'-Flap structure.
[0075] To demonstrate the importance of un...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com