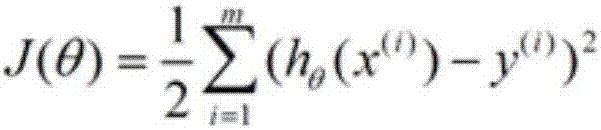Method of analysis of regulatory relationship between genes or loci and disease based on multi-omics
A locus and gene technology, applied in the field of multi-omics analysis of the relationship between genes or loci and disease regulation, can solve problems such as large random interference, inaccurate analysis results, and different environmental effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0011] The present invention will be described in further detail below in conjunction with specific embodiments, but the embodiments of the present invention are not limited thereto.
[0012] Collect patient sample data, in which the condition and etiology of these patients are known. For example, it is known that albinism is mainly caused by the lack of normal tyrosinase gene, which leads to the inability to convert tyrosine into melanin. Phenylketonuria is mainly caused by Inability to convert phenylalanine to tyrosine due to lack of normal gene for phenylalanine hydroxylase. These data are used as the training set, and the linear regression method is used to fit the sample data to train a suitable model.
[0013] The specific methods and principles are as follows:
[0014] The collected sample data were analyzed using transcriptome analysis, CHIP-Seq analysis, miRNA analysis and GWAS analysis respectively. For the same patient, the analysis results were obtained separatel...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com