Universal molecular marker primer for tree and joint development method and application thereof
A technology of molecular markers and trees, which is applied in biochemical equipment and methods, microbiological determination/inspection, DNA/RNA fragments, etc., can solve problems such as the inability to extract intron sequences, achieve objective results and improve accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
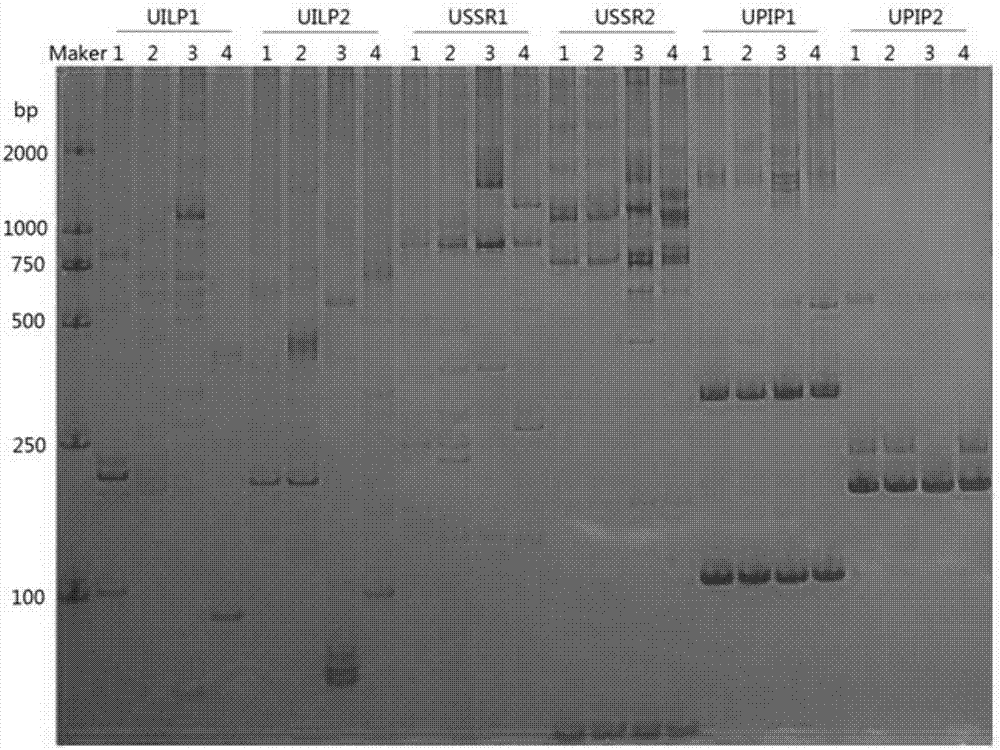
[0033] The present invention will be specifically introduced below in conjunction with the accompanying drawings and specific embodiments.
[0034] refer to figure 1 , the joint development method of the SSR of tree of the present invention, ILP and PIP universal molecular marker primer, comprises the following steps:
[0035] Step1: SSR site and intron site scanning
[0036] Download all tree genome sequences and genome annotation information from the public data platform, scan the SSR site, intron site and predicted intron site in the genome sequence, and obtain the corresponding site information.
[0037] It is generally believed that the minimum length of an SSR site is 12bp, that is, a single nucleotide repeat unit is at least 12 repeats, a dinucleotide repeat unit is at least 6 repeats, a trinucleotide repeat unit is at least 4 repeats, and a tetranucleotide repeat is at least 4 repeats. The unit is at least 3 repeats, the pentanucleotide repeating unit is at least 3 r...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com