Duck circovirus ORF3 nucleus location NLS sequence and application thereof
A nuclear localization signal and sequence technology, which is applied in the field of duck circovirus ORF3 protein nuclear localization NLS sequence, can solve problems such as homology differences of ORF3 nucleic acid sequences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1 The C-terminal differential region of ORF3 was mutated to determine the precise NLS sequence: RRLRTCNCRACRTLKH.
[0030] 1 Materials and methods
[0031] 1.1 Plasmid
[0032] The pEGFP-N3, pcDNA-FJ-ORF3 and pcDNA-WF-ORF3 plasmids are commercially available or prepared by the laboratory. The plasmids are transformed into Escherichia coli strain DH5α, the plasmid DNA is extracted, and the DNA concentration is measured with a DNA / RNA quantifier, and placed in - Store at 20°C for later use.
[0033] 1.2 Main reagents
[0034] Pfu Turbo DNA Polymerase was purchased from Agilent; DpnI was purchased from NEB; Lipo2000 was purchased from Invitrogen.
[0035] 1.3 Location analysis of pEGFP-FJ-ORF3 and pEGFP-WF-ORF3
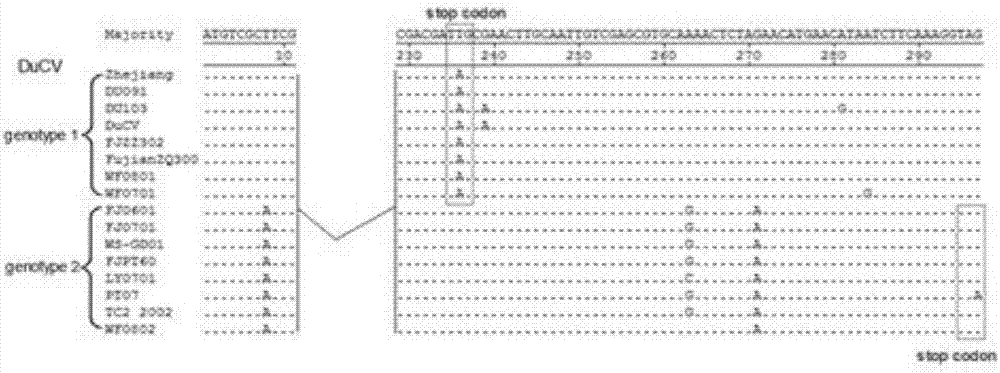
[0036] 1.3.1 ORF3 sequence analysis of genotype I and II DuCV
[0037] Compared with type I and type II DuCV ORF3 gene sequences, the 236th nucleotide of ORF3 of type I strains undergoes a T→A mutation, and the codon TTG (235-237) is converted into a ...
Embodiment 2
[0075] Example 2 Fusion expression of the NLS signal in the C-terminal differential region with GFP protein, and the ability of EGFP to obtain nuclear localization
[0076] 2.1 Construction of pNLS-GFP expression vector and its expression measurement
[0077] pNLS-GFP plasmid construction: synthesize the following two primers, the slash mark is the NLS sequence
[0078] Primer NLS-F1:
[0079] AATTCATGCGACGATTGCGAACTTGCAATTGTCGAGCGTGCAGAACTCTAAAACATG
[0080] Primer NLS-R1:
[0081] GATCCATGTTTTAGAGTTCTGCACGCTCGACAATTGCAAGTTCGCAATCGTCGCATG
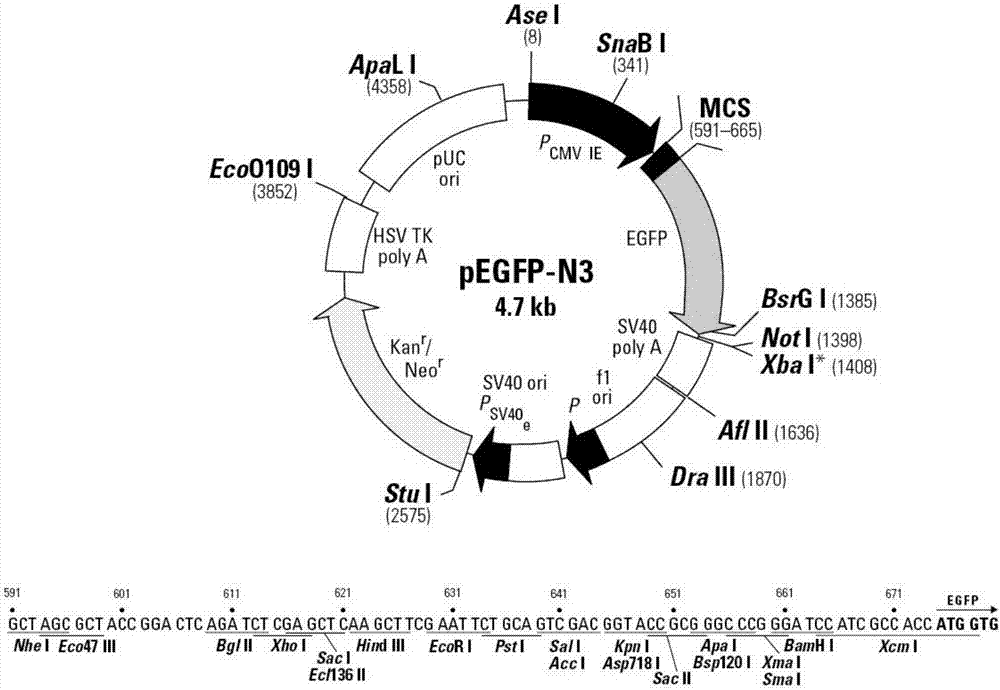
[0082] Anneal the upstream and downstream, and clone the NLS into pEGFP-N3 using the EcoRI and BamHI restriction sites (plasmid map as shown in figure 1 Shown) in the carrier, the specific test steps are carried out according to routine tests.
[0083] 2.2 NLS-GFP localization analysis
[0084] 2.2.1 Fluorescence test
[0085] 1. Place the flyer in the culture plate
[0086] 2. DF-1 cells were subcultured in 6-well cell culture pla...
Embodiment 3
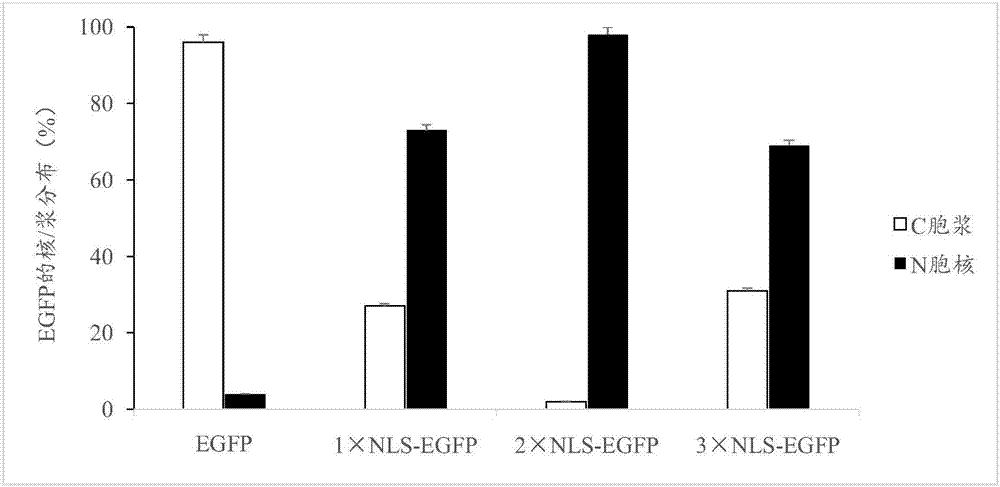
[0103] Example 3 Nuclear Localization Effects of Different Copy Number NLS Sequences
[0104] 3.1 Construction of p2*NLS-GFP and p3*NLS-GFP expression vectors
[0105] Synthesize the following two sets of primers, the slash mark is the NLS sequence
[0106] Primer NLS-F2:
[0107] AATTCATGCGACGATTGCGAACTTGCAATTGTCGAGCGTGCAGAACTCTAAAACATATGCGACGATTGCGAACTTGCAATTGTCGAGCGTGCAGAACTCTAAAACATG
[0108] Primer NLS-R2:
[0109] GATCCATGTTTTAGAGTTCTGCACGCTCGACAATTGCAAGTTCGCAATCGTCGCATATGTTTTGAGTTCTGCACGCTCGACAATTGCAAGTTCGCAATCGTCGCATG
[0110] Primer NLS-F3:
[0111] AATTCATGCGACGATTGCGAACTTGCAATTGTCGAGCGTGCAGAACTCTAAAACATATGCGACGATTGCGAACTTGCAATTGTCGAGCGTGCAGAACTCTAAAACATATGCGACGATTGCGAACTTGCAATTGTCGAGCGTGCAGAACTCTAAAACATG
[0112] Primer NLS-R3:
[0113] GATCCATGTTTTAGAGTTCTGCACGCTCGACAATTGCAAGTTCGCAATCGTCGCATATGTTTTAGAGTTCTGCACGCTCGACAATTGCAAGTTCGCAATCGTCGCATATGTTTTAGAGTTCTGCACGCTCGACAATTGCAAGTTCGCAATCGTCGCATG
[0114] Anneal the upstream and downstream, and clone the NLS int...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com