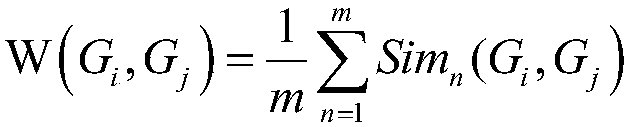A method for constructing regulatory networks of differentially expressed genes in specific cancers
A cancer differential expression and gene regulation network technology, applied in special data processing applications, instruments, calculations, etc., can solve the problems of reducing computational complexity, single data source, and high computational complexity, so as to reduce complexity and improve computational complexity. Precision, high-precision effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0038] The present invention will be further described in detail below in conjunction with the accompanying drawings, so that those skilled in the art can implement it with reference to the description.
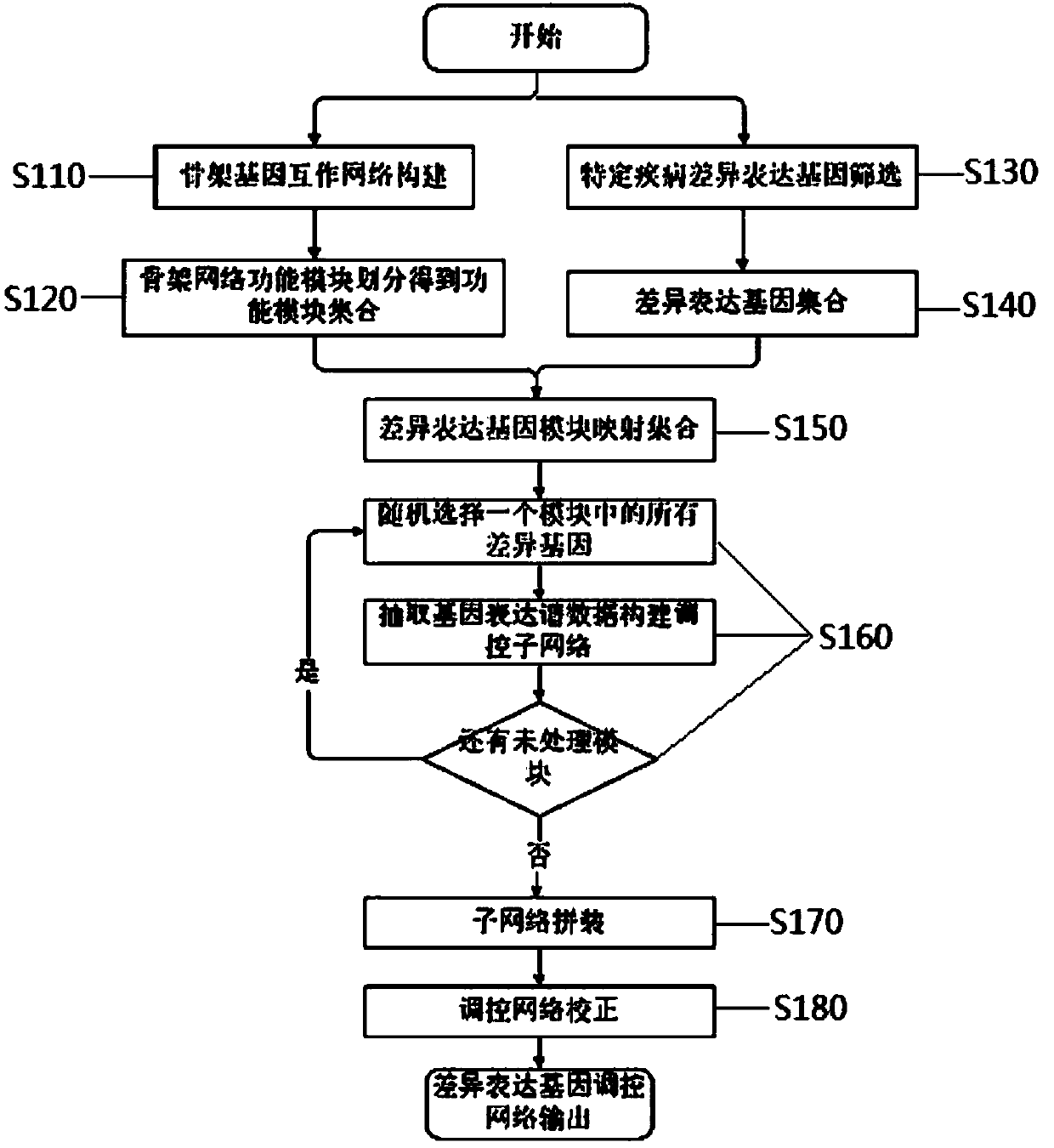
[0039] Such as figure 1 As shown, the present invention provides a method for constructing a specific cancer differentially expressed gene regulatory network, and the specific steps are as follows:
[0040] Step 1 S110, construction of backbone gene interaction network.
[0041] The skeleton gene interaction network is a weighted network, in which the weight represents the functional similarity between genes, and the weight calculation method is as follows:
[0042]
[0043]Among them, m represents the number of types of gene-related information. In this method, m=3 is preferred, and only three aspects of information, namely gene ontology annotation information, gene metabolic pathway information, and known inter-gene interaction relations, are considered. Sim n (G i ,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com