Preparation method of gene-encoded whole protein catenane
A protein and catenane technology, applied in the direction of animal/human protein, p53 protein, mammalian protein, etc., can solve the problem of not preparing molecular weight recombinant protein catenane, and achieve the effect of reducing difficulty
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1: Construction of AEXEB and EAXEB fusion protein plasmids and expression in Escherichia coli
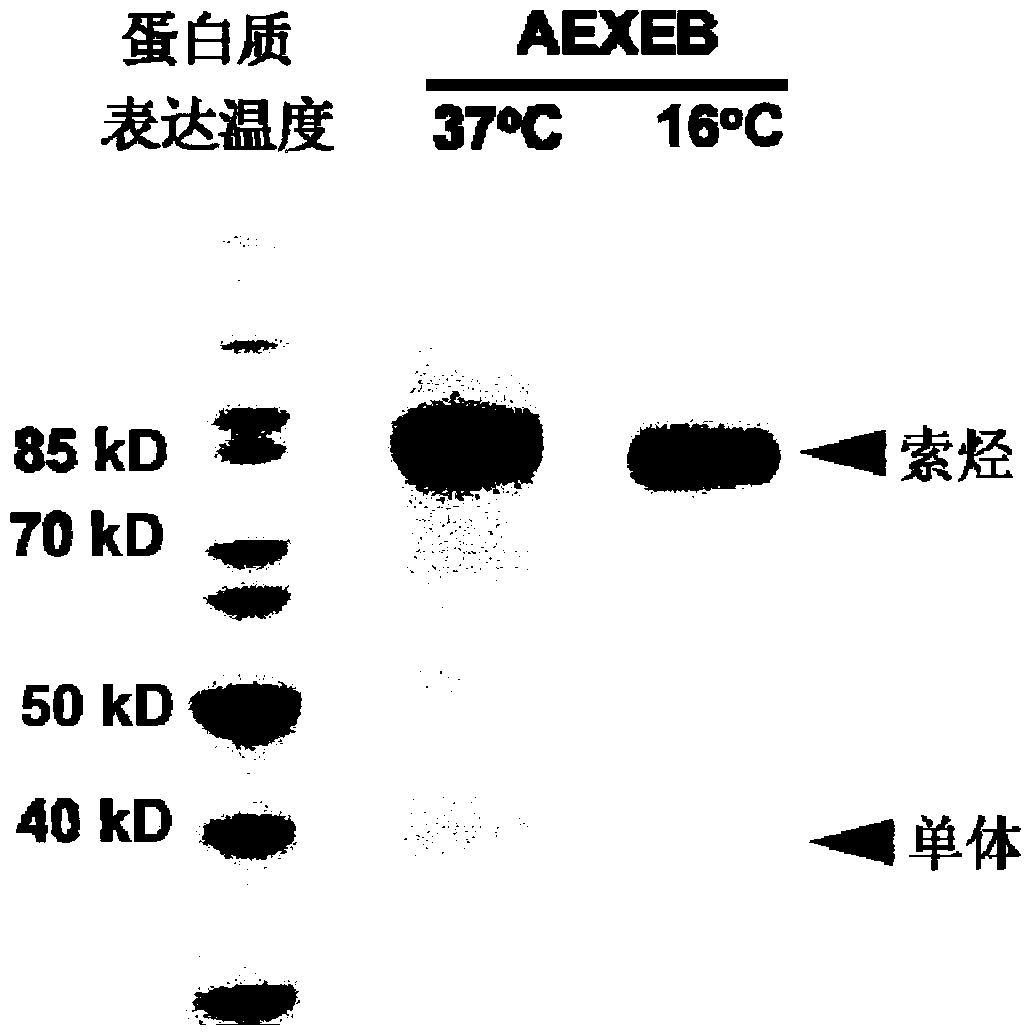
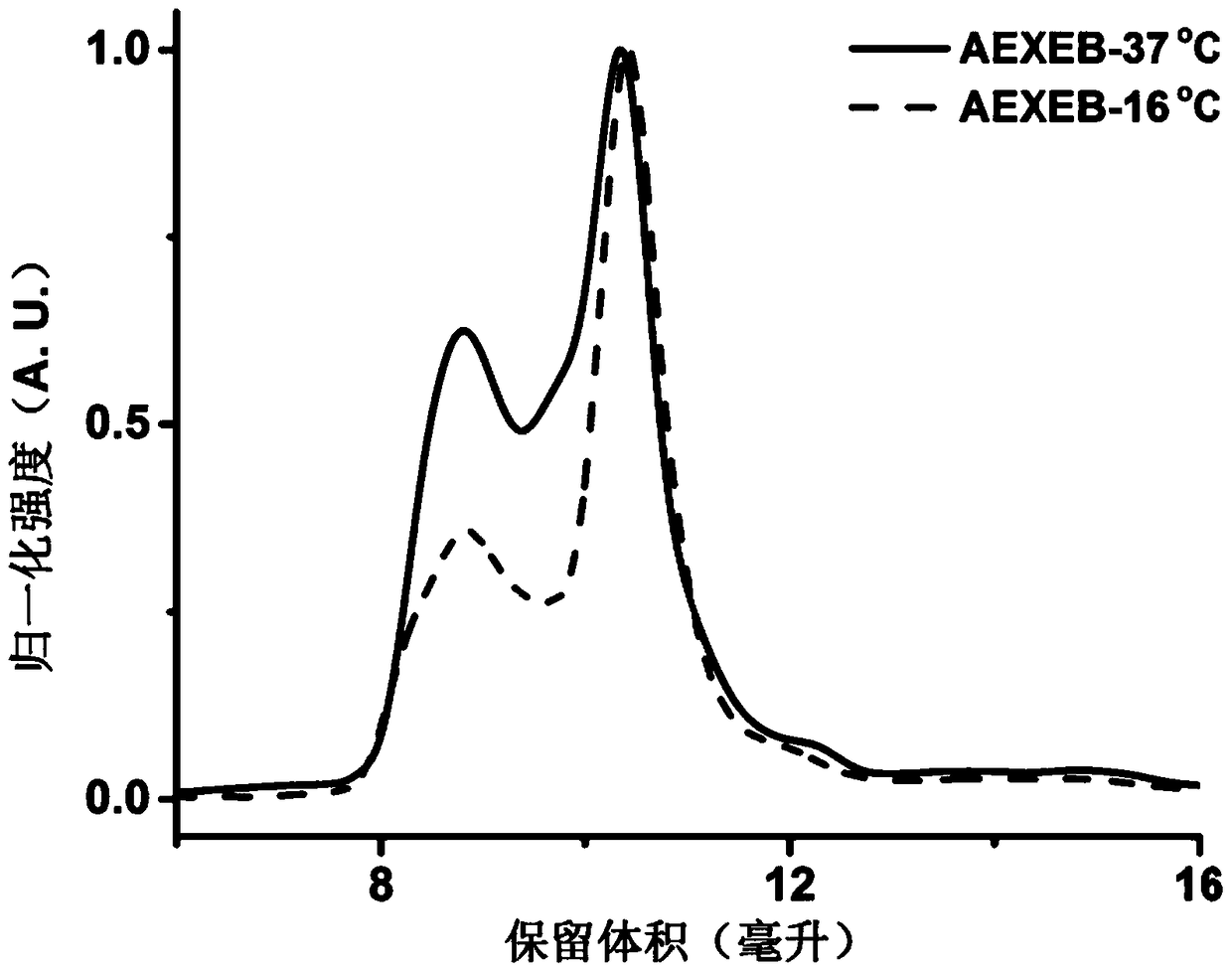
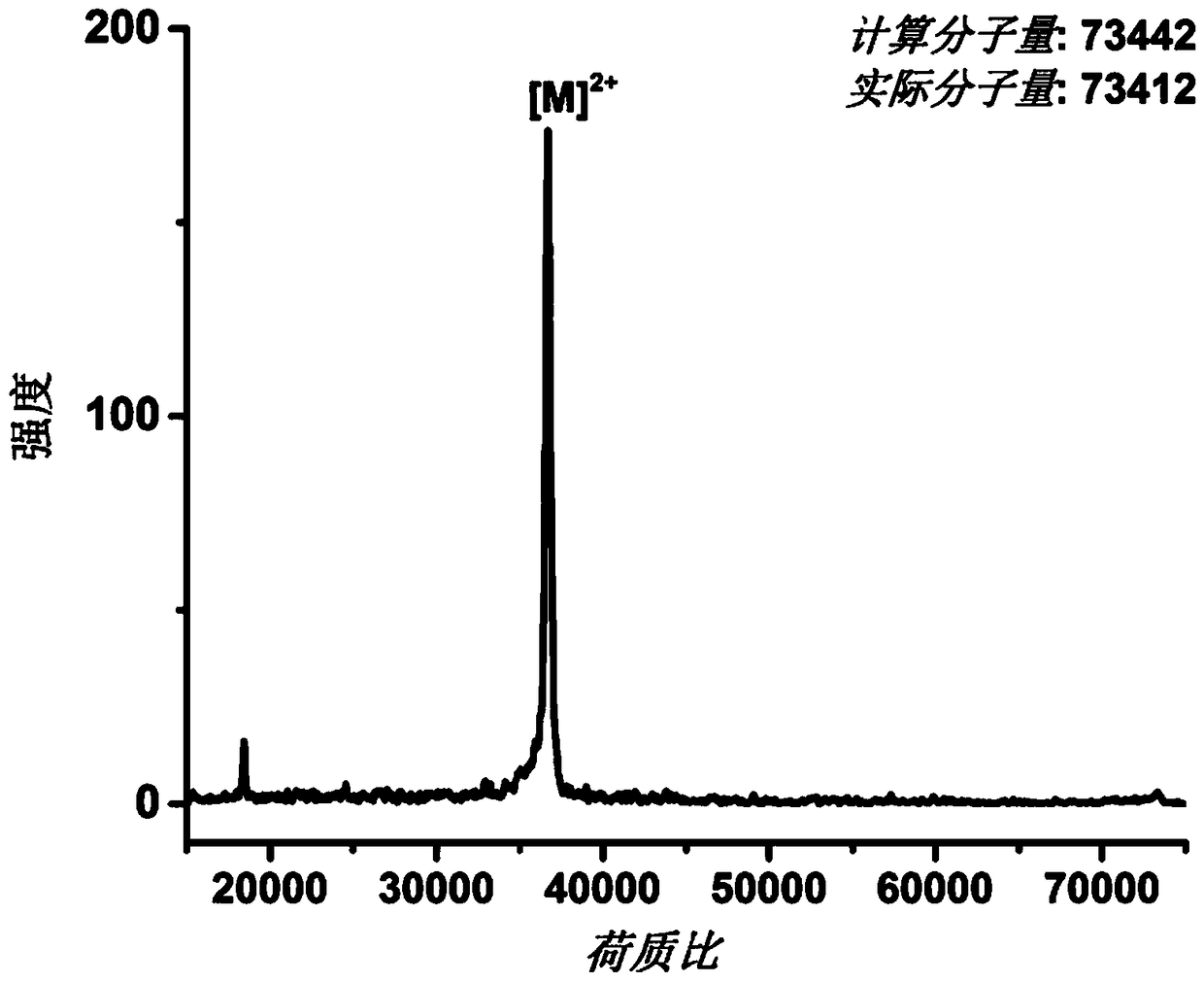
[0043] The AEXEB sequence constructed in this example is shown as SEQ ID No: 7 in the sequence table, wherein the 14th to 26th positions are A, the 29th to 108th positions and 151 to 230 are E, the 111th to 148th positions are X, and the 111th to 148th positions are X. 233-359 are B, and 237-243 are TEV restriction sites.
[0044] The EAXEB sequence constructed in this example is shown as SEQ ID No: 8 in the sequence table, wherein the 96th to 108th positions are A, the 14th to 93rd positions and 154 to 233 are E, the 114th to 151st positions are X, and the 114th to 151st positions are X. 238-364 are B, and 242-248 are TEV restriction sites.
[0045] The gene sequence of the above fusion protein was introduced into the vector pQE-80L, and then transformed into Escherichia coli.
[0046] Expressed in E. coli BL21(DE3). Before large-scale expression, the transformed ...
Embodiment 2
[0047] Example 2: Construction of AXB and BXA fusion protein plasmids and expression in Escherichia coli
[0048] The AXB sequence constructed in this example is shown as SEQ ID No: 9 in the sequence table, wherein the 16th to 28th are A, the 33rd to 70th are X, the 77th to 203rd are B, and the 81st to 87th are TEV restriction site.
[0049] The BXA sequence constructed in this example is shown as SEQ ID No: 10 in the sequence listing, where the 17th to 143rd is B, the 21st to 27th is the TEV restriction site, the 156th to 193rd is X, and the 199th ~211 bits are A.
[0050] The above protein sequence is constructed in the pQE-80L expression vector. Subsequently expressed in E. coli BL21(DE3). Before large-scale expression, the transformed monoclonal bacteria were first inoculated in 5 mL 2×YT medium (containing 100 μg / mL ampicillin), cultured overnight at 37°C with shaking at 250 rpm, and transferred to In 1L of fresh 2×YT medium (in a 3L Erlenmeyer flask, containing 100μg...
Embodiment 3
[0051] Example 3: Purification of AEXEB and EAXEB Catalane Proteins by Nickel Affinity Chromatography Column
[0052] Collect 1L of Escherichia coli culture solution in a centrifuge bottle, collect the bacteria by centrifugation at 5000g, and remove the supernatant culture solution. Resuspend the bacteria with 40mL denaturing buffer solution A (20mM disodium hydrogen phosphate, 500mM sodium chloride, 25mM imidazole, 8M urea, pH=8.0) containing 2% Tween 20 and 1mM phenylmethylsulfonyl fluoride, and then use The sonicator disrupted the cells at 4°C, and then centrifuged the Escherichia coli crushed product at 4°C under a centrifugal force of 28000g for 20 minutes. The resulting supernatant was mixed with the nickel-affinity resin equilibrated with denaturing buffer solution A, and stirred at 4°C for 1 h. The mixture was placed in an empty column and eluted by gravity flow. The eluent was denaturing buffer solution B (20 mM disodium hydrogen phosphate, 500 mM sodium chloride, 8 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com