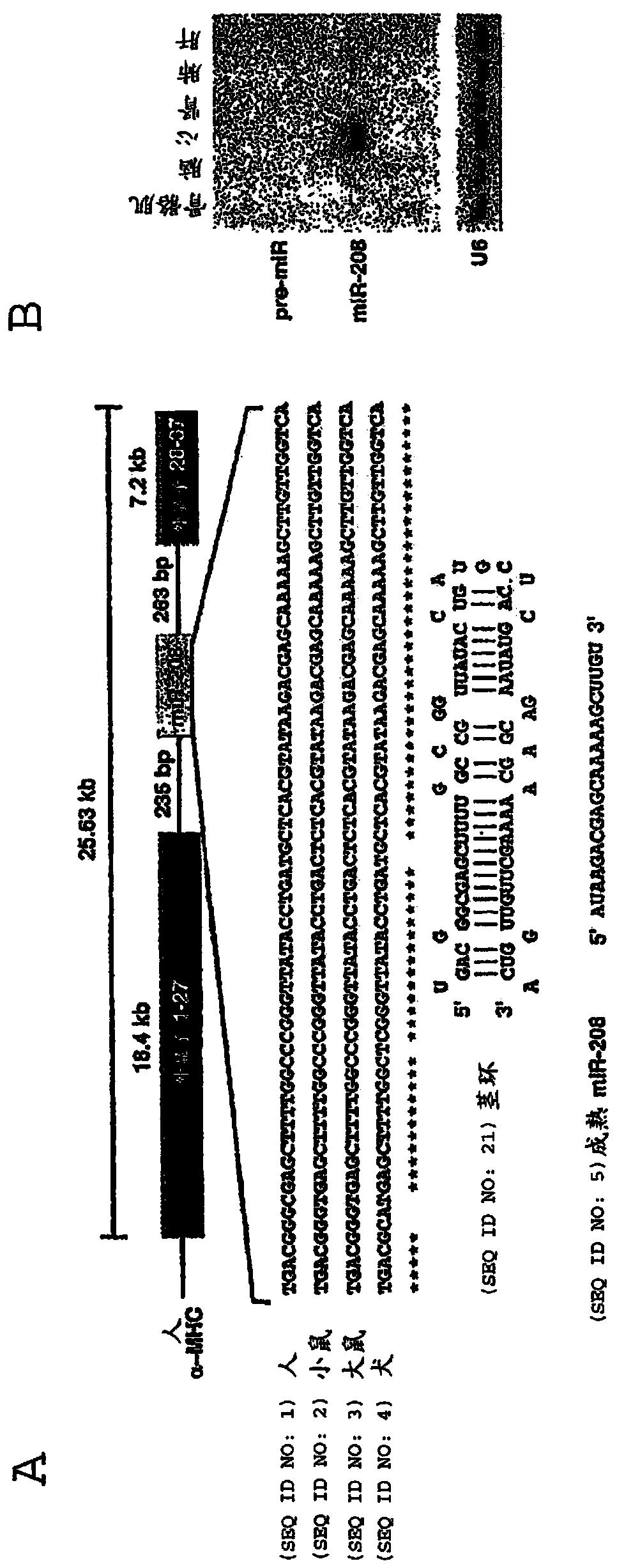
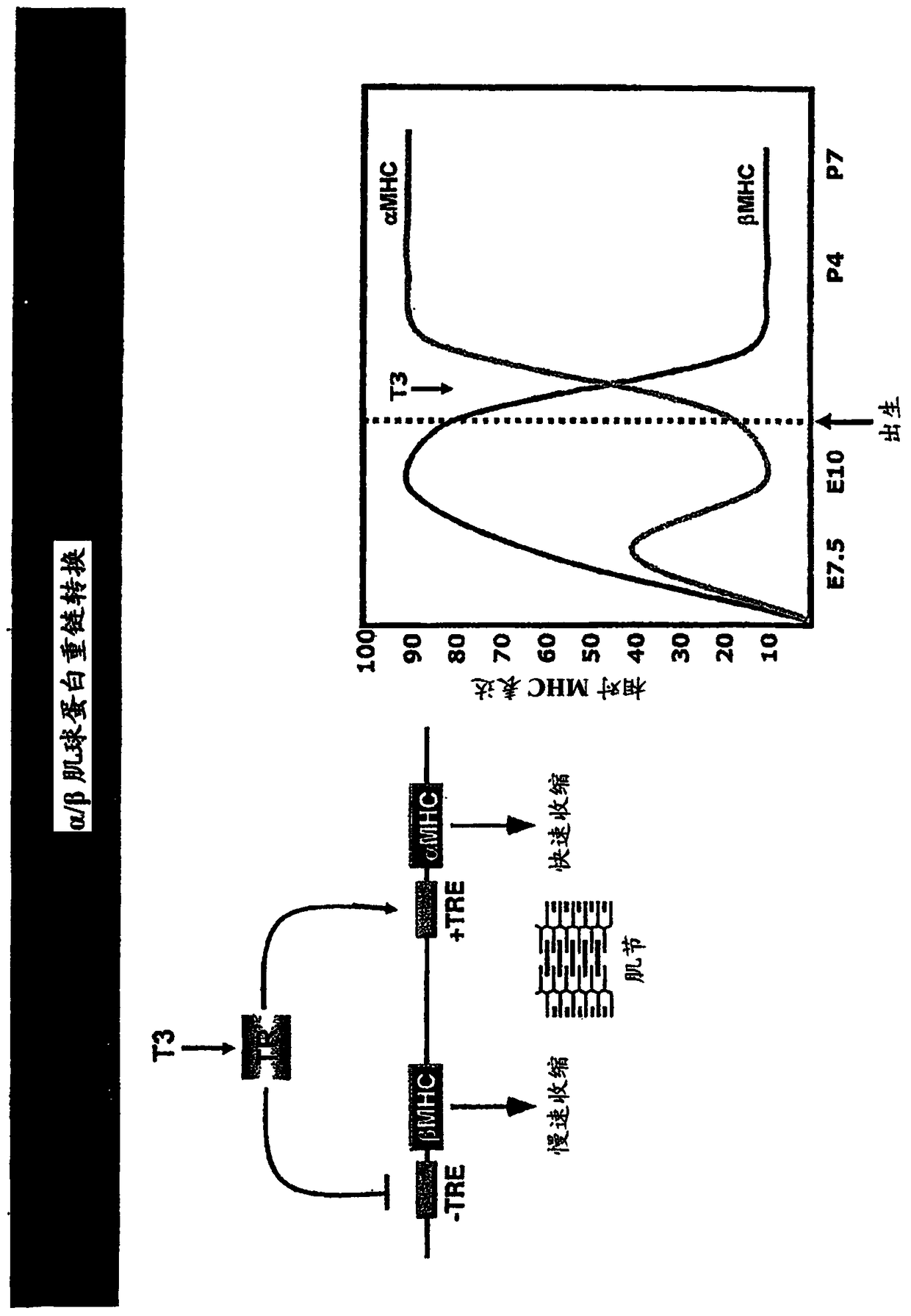
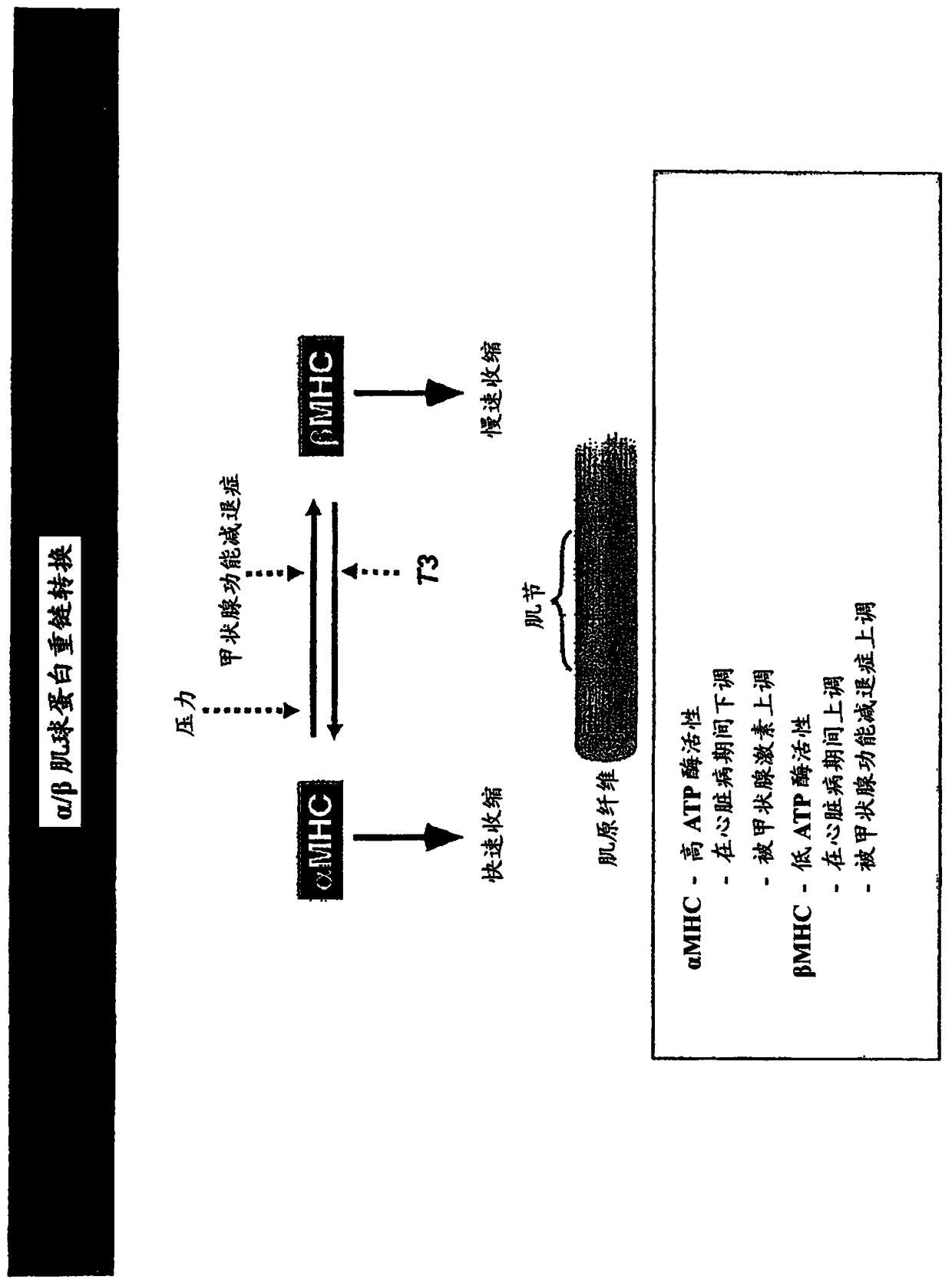
Family of microRNAs regulating fibrosis and their uses
A technique for tissue fibrosis, application in developmental biology and molecular biology, capable of solving complex problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0394] Example 1: Regulation of Cardiac Hypertrophy and Heart Failure by Pressure Responsive miRNAs
[0395] Based on their involvement in regulating cellular phenotypes, the inventors hypothesized that miRNAs play a role in regulating the cardiac response to cardiac stress, which is known to result in transcriptional and translational changes in gene expression. To investigate the possible involvement of miRNAs in cardiac hypertrophy, they performed a side-by-side miRNA Microarray analysis. Mice (TAB) (Hill et al., 2000) subjected to thoracic aortic bandage (which induces hypertrophy by increasing cardiac afterload) were compared with sham-operated animals. In a second model, transgenic mice (Molkentin et al., 1998) expressing activated calcineurin (CnA) in the heart (which leads to a severe, characterized form of hypertrophy) were compared with wild-type littermates A comparison was made (FIG. 14A). RNA isolated from hearts of TAB mice showed elevated expression of 27 miR...
Embodiment 2
[0397] Example 2: Discovery of the miR-29 family as downstream targets of miR-208 regulation
[0398] In an effort to identify downstream miRNAs that might mediate the effects of miR-208, the inventors performed miRNA microarrays on hearts from wild-type and miR-208-null mice ( Figure 16 ). They found that multiple members of the miR-29 family were upregulated in miR-208 null mice ( Figure 17 ). Target predictions indicate that miR-29 family members target mRNAs encoding various collagens and other components of the extracellular matrix ( Figure 18 ). Thus, upregulation of miR-29 family members in miR-208-null mice may account for the blockade of fibrosis seen in these animals ( Figure 19 ).
[0399] The discovery that miR-29a-c is downregulated and targets mRNAs encoding collagen and extracellular matrix proteins in diseased hearts suggests that strategies to enhance miR-29a-c expression or its binding to target mRNAs may contribute to pathological cardiac remodeling...
Embodiment 3
[0400] Example 3: miR-29a-c regulates the expression of fibrotic genes
[0401] To begin to define the possible functions of miR-29a-c in the post-MI heart, the inventors used computational predictions to identify possible targets of miR-29a-c. The Targetscan predictions web site points to an unexpectedly high number of fibrosis-associated mRNAs encoding collagens, metallopeptidases, and integrins as possible miR-29a-c targets (web site at targetscan.org). To determine whether miR-29a-c downregulation might regulate cardiac fibrosis, the inventors focused on predicted targets involved in ECM generation in the heart. Elastin (ELN), fibrillin 1 (FBN1), type I collagen α1 and α2 (COL1A1, COL1A2), and type III collagen α1 (COL3A1) all contain one or more conserved potential seeds of miR-29a-c sequence (FIG. 20A).
[0402]Because miRNAs downregulate the steady-state levels of their target mRNAs as well as translation, the inventors analyzed the expression of predicted miR-29a-cmR...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com