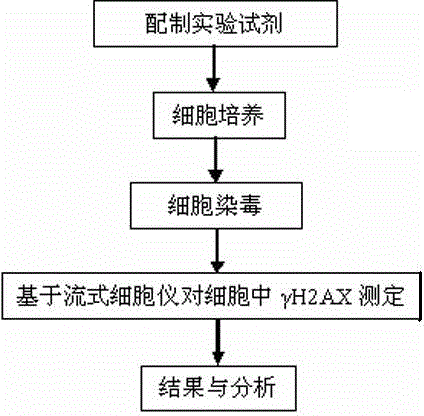
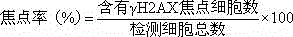Gamma H2AX based cell DNA damage detecting method
A DNA damage and H2AX technology, applied in measurement devices, biological tests, material inspection products, etc., can solve the problems of large loss of cells after repeated washing, non-specific fluorescence interference, complicated operation steps, etc., and achieve fast and convenient detection process. The operation method is simple and the effect of improving the detection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0027] Example 1 Evaluation of NNK
[0028] Collect CHO cells in the exponential growth phase at 1×10 6 Each cell / well was seeded in a 6-well plate, cultured in a cell incubator for 24 hours, and then exposed to NNK for 24 hours. In the experiment, the doses of NNK were 0 μg / mL, 25 μg / mL, 50 μg / mL, 100 μg / mL, 200 μg / mL and 400 μg / mL to infect CHO cells, and three parallel wells were set up for each condition , placed at 37 °C, 5% CO 2 cultured in a cell culture incubator for 24 h. After the cells were infected, add 500 μL of 0.25% trypsin, digest for 1-2 min, discard the trypsin, add 1 mL of phosphate buffered saline (PBS) to suspend, centrifuge at 1500 rpm / min, discard the supernatant, Wash twice. Ethanol fixation: Prepare 70% ethanol and pre-cool at -20°C. After vortexing to loosen the cell aggregates, 1 mL of pre-cooled 70% ethanol was added drop by drop while vortexing, and placed in a -20 °C refrigerator for 24 h. TritonX-100 membrane rupture: Take out the fixed sam...
example 2
[0031] Example 2 Evaluation of AαC
[0032] Collect CHO cells in the exponential growth phase at 1×10 6 Each cell / well was seeded in a 6-well plate, cultured in a cell incubator for 24 h, and then AαC was exposed for 24 h. In the experiment, the doses of AαC were 0 μg / mL, 2.5 μg / mL, 5 μg / mL, 10 μg / mL, 20 μg / mL and 40 μg / mL to infect CHO cells, and three parallel wells were set up for each condition , placed at 37 °C, 5% CO 2 cultured in a cell culture incubator for 24 h. After the cells were infected, add 500 μL of 0.25% trypsin, digest for 1-2 min, discard the trypsin, add 1 mL of phosphate buffered saline (PBS) to suspend, centrifuge at 1500 rpm / min, discard the supernatant, Wash twice. Ethanol fixation: Prepare 70% ethanol and pre-cool at -20°C. After vortexing to loosen the cell aggregates, add 1 mL of pre-cooled 70% ethanol drop by drop while vortexing, and place in a -20 °C refrigerator overnight for fixation. TritonX-100 membrane rupture: Take out the fixed sample...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com