Molecular marker for identifying carp varieties and method for identifying by utilizing molecular marker
A molecular marker and variety technology, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve problems such as inability to accurately identify carp species
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
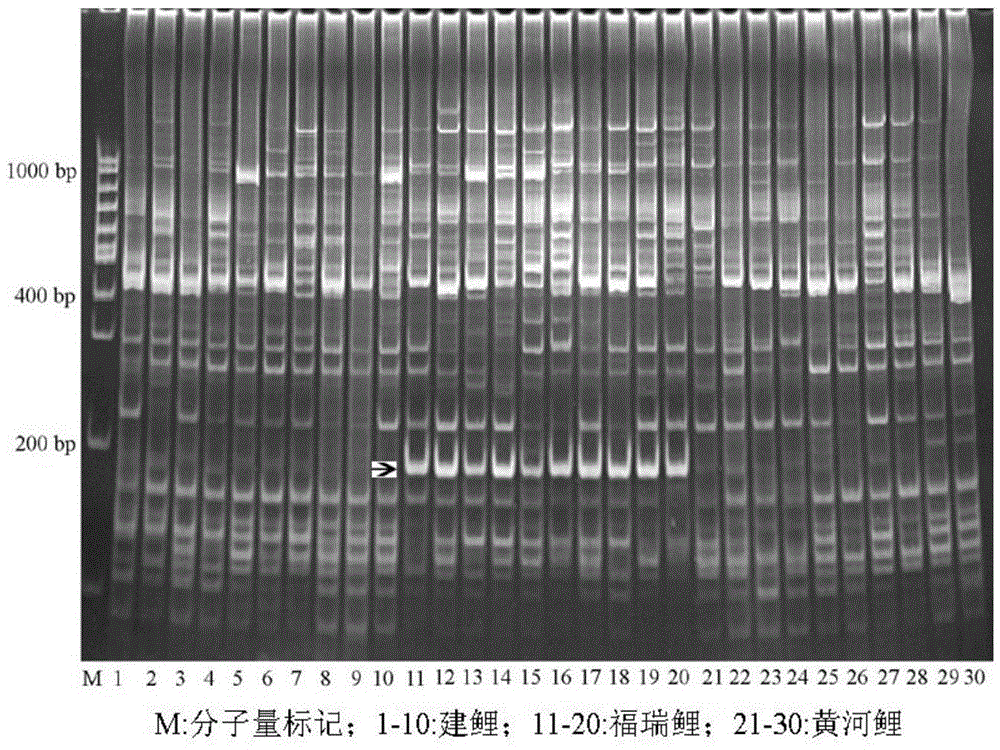
[0018] Furui carp and its original parents—Jian carp and Huanghe carp were obtained from the Yixing Experimental Base of the Freshwater Fishery Research Center of the Chinese Academy of Fishery Sciences, and 10 of each fish were randomly selected. Genomic DNA was extracted from the fin rays of each fish.
[0019] GH(GenBank accession no.M27000.1), GHR(AY741100.1), IGF(D83272.1), TRH(AB179818.1) and GTH(X56497.1) genes that may be related to carp growth traits were selected as target candidates Gene, 18 fixed primers were designed. According to the characteristics of exons or introns, four random primers rich in GC or AT core regions were designed and synthesized according to Hu (2006). The details are shown in Table 1.
[0020] Using the genomic DNA as a template, 18 fixed primers were combined with 4 random primers for PCR amplification. The primers are shown in Table 1.
[0021] The total volume of the PCR reaction was 15 μL, where Mg 2+ 1.5mmol / L, dNTPs0.35mmol / L, rando...
Embodiment 2
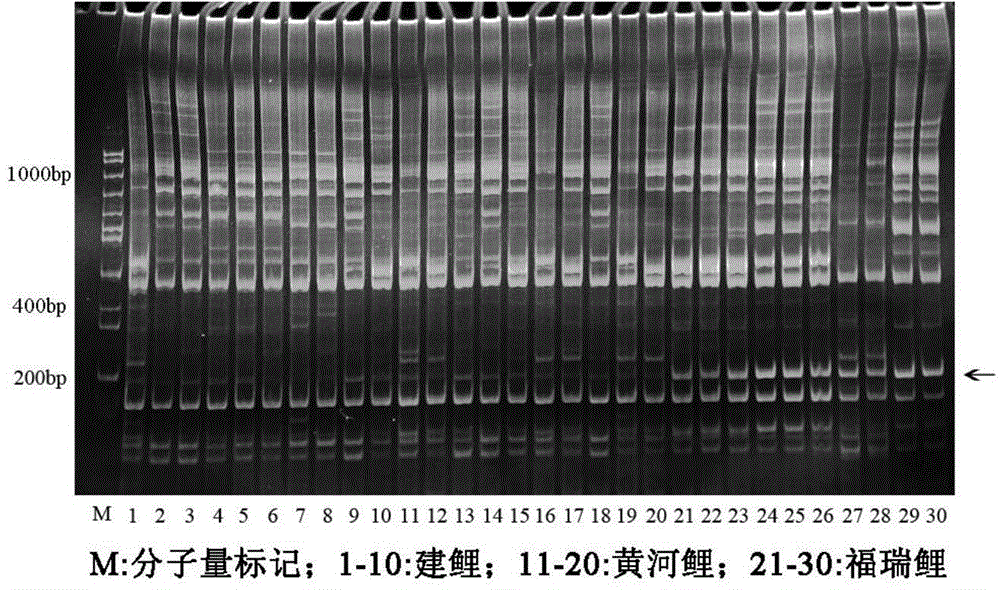
[0025] Furui carp and one of its original parents, Jian carp, was taken from the Nanquan Experimental Base of Freshwater Fishery Research Center of the Chinese Academy of Fishery Sciences, and the other original parent, the Yellow River carp, was taken from the Yellow River Carp Breeding Farm in Zhengzhou, Henan. Take 10 tails. Genomic DNA was extracted from the fin rays of each fish.
[0026] Using the genomic DNA as a template, 18 fixed primers were combined with 4 random primers for PCR amplification. The primers are shown in Table 1.
[0027] The total volume of the PCR reaction was 15 μL, where Mg 2+ 1.5mmol / L, dNTPs0.35mmol / L, random primer 6pmol / L, fixed primer 10pmol / L, DNA template 60ng, Taq DNA polymerase 1.0U, make up the volume with sterilized double distilled water. The PCR reaction conditions were: 94°C for 4min; 94°C for 45s, 35°C for 45s, 72°C for 1min, 5 cycles; 94°C for 45s, 53°C for 45s, 72°C for 1min, 35 cycles; 72°C for 10min.
[0028] After the PCR rea...
Embodiment 3
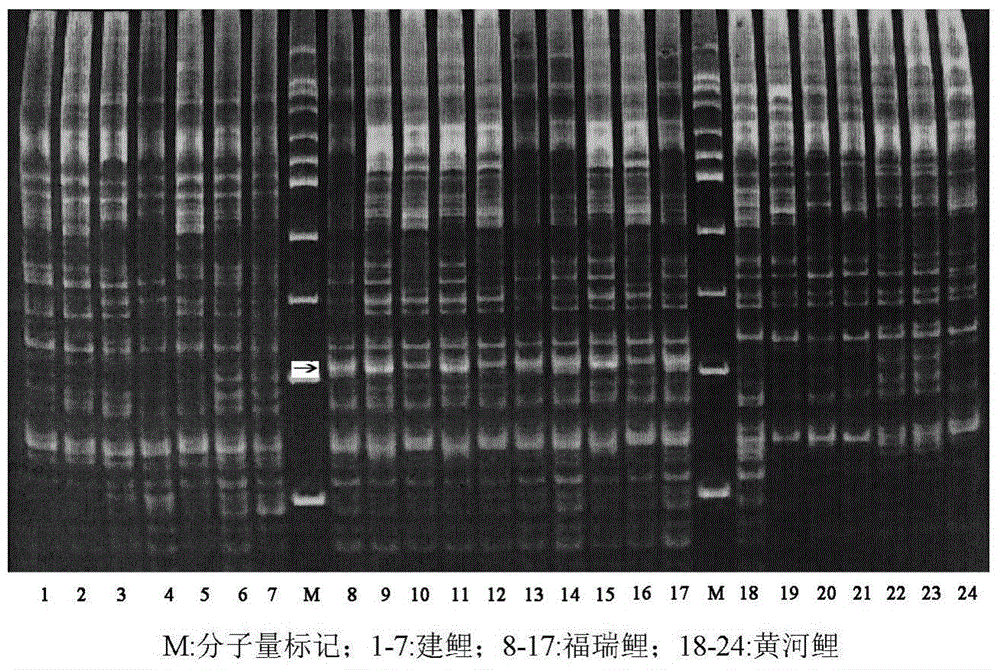
[0031] Ten Furui carp and their original parents—7 Jian carp and 7 Huanghe carp were obtained from the Yangtze River Fisheries Research Institute of the Chinese Academy of Fishery Sciences. Genomic DNA was extracted from the fin rays of each fish.
[0032] Using the genomic DNA as a template, 18 fixed primers were combined with 4 random primers for PCR amplification. The primers are shown in Table 1.
[0033] The total volume of the PCR reaction is 15 μl, where Mg 2+ 1.5mmol / L, dNTPs0.35mmol / L, random primer 6pmol / L, fixed primer 10pmol / L, DNA template 60ng, Taq DNA polymerase 1.0U, make up the volume with sterilized double distilled water. The PCR reaction conditions were: 94°C for 4min; 94°C for 45s, 35°C for 45s, 72°C for 1min, 5 cycles; 94°C for 45s, 53°C for 45s, 72°C for 1min, 35 cycles; 72°C for 10min.
[0034] After the PCR reaction, the products were separated by electrophoresis using 8.0% non-denaturing polyacrylamide gel, and after staining by silver staining, pho...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com