Fluorescence activated cell sorting (FACS) enrichment to generate plants
A plant cell, plant technology, applied in the field of fluorescence activated cell sorting
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
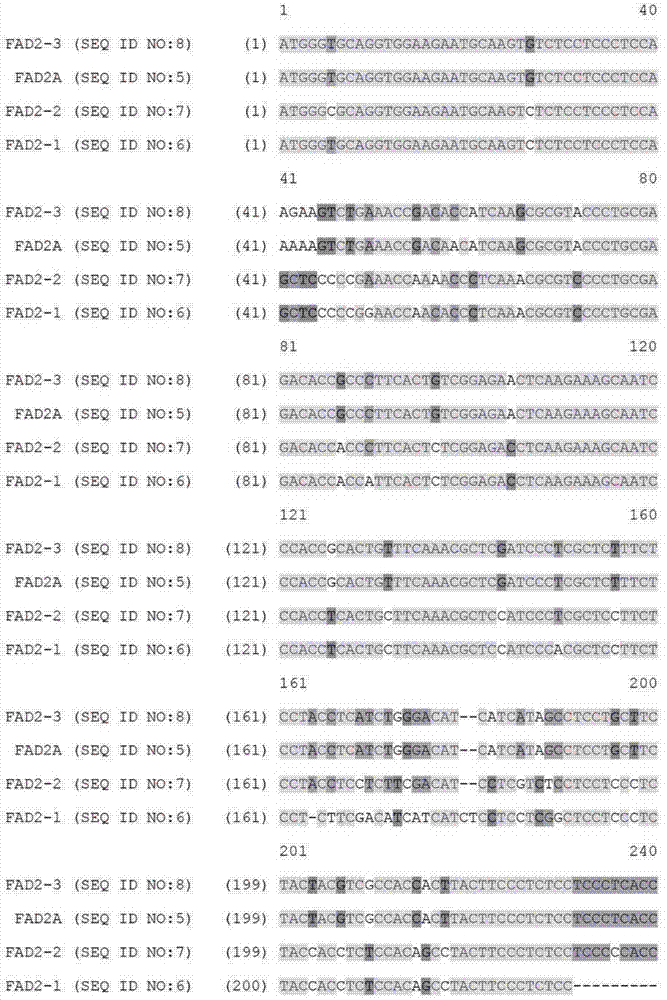
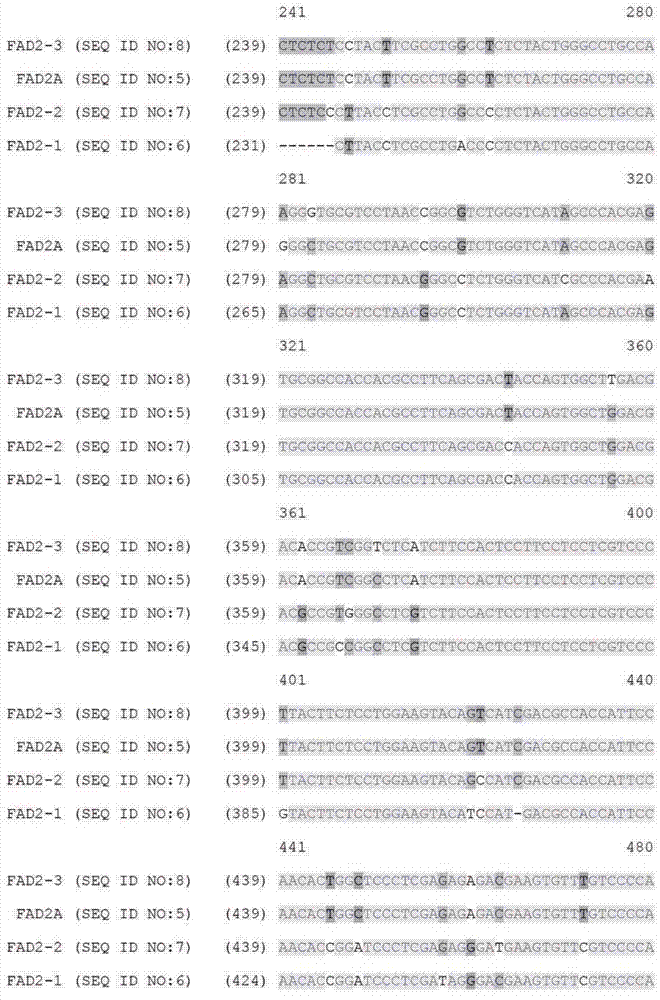
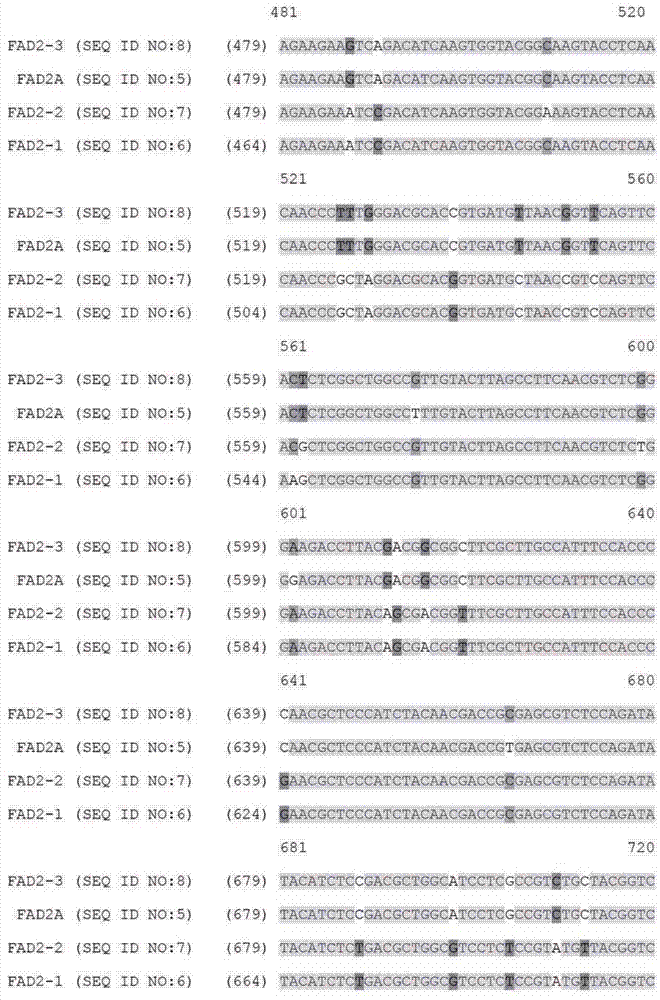
[0089] Example 1: Identification of paralogous FAD2 and FAD3 target sequences from bacterial artificial chromosome libraries
[0090] BAC construction
[0091] Bacterial artificial chromosome (BAC) libraries were purchased from a supplier (Amplicon Express, Pullman, WA). The BAC library consisted of 110,592 BAC clones containing high molecular weight genomic DNA (gDNA) fragments isolated from Brassica napus var. DH10275. Digest gDNA with BamHI or HindIII restriction enzymes. The isolated gDNA fragment of approximately 135 Kbp was ligated into pCC1BAC vector (Epicentre, Madison, WI) and transformed into E. coli strain DH10B (Invitrogen). The BAC library consisted of an even number of BAC clones constructed with two different restriction enzymes. Therefore, the BAC library constructed by HindIII consisted of 144 384-well plates. Similarly, the BAC library constructed by BamHI consisted of 144 384-well plates. A total of 110,592 BAC clones were isolated and arrayed into 28...
Embodiment 2
[0123] Example 2: Design of a Zinc Finger Binding Domain Specific to the FAD2 Gene
[0124]Zinc finger proteins were designed against DNA sequences encoding various functional sequences of the FAD2 gene locus as previously described. See, eg, Urnov et al. (2005) Nature 435:646-651. Exemplary target sequences and recognition helices are shown in Tables 6 and 8 (recognition helix region design) and Tables 7 and 9 (target sites). In Tables 8 and 9, nucleotides in the target site that are in contact with the ZFP recognition helix are indicated in capital letters; non-contacting nucleotides are indicated in lower case. Zinc finger nuclease (ZFN) target sites were designed to bind 5 target sites of FAD2A and 7 target sites of FAD3. The FAD2 and FAD3 zinc fingers were designed to be incorporated into a zinc finger expression vector encoding a protein having at least one finger with a CCHC structure. See US Patent Publication No. 2008 / 0182332. In particular, the last finger in eac...
Embodiment 3
[0141] Example 3: Evaluation of Zinc Finger Nuclease Cleavage of the FAD2 Gene
[0142] construct assembly
[0143] Plasmid vectors containing ZFN expression constructs for exemplary zinc finger nucleases identified using yeast assays as described in Example 2 were designed and completed using skills and techniques generally known in the art. Each zinc finger coding sequence was fused to a sequence encoding the opaque-2 nuclear localization signal (Maddaloni et al. (1989) Nuc. Acids Res. 17(18):7532), upstream of the zinc finger nuclease.
[0144] Next, the opaque-2 nuclear localization signal::zinc finger nuclease fusion sequence is paired with the complementary opaque-2 nuclear localization signal::zinc finger nuclease fusion sequence. Thus, each construct consisted of a single open reading frame consisting of the 2A sequence from the genus spp. (beta tetrasomy) virus (Mattion et al. (1996) J. Virol. 70:8124- 8127) separated by two opaque-2 nuclear localization signal::z...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com