Method for sequencing high-flux circular RNA (ribonucleic acid)
A high-throughput, sequencing technology, applied in the field of molecular biology, can solve the problem of inaccurate sequencing of circular RNA, and achieve the effects of reducing the amount of sequencing data, improving efficiency, and increasing enrichment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
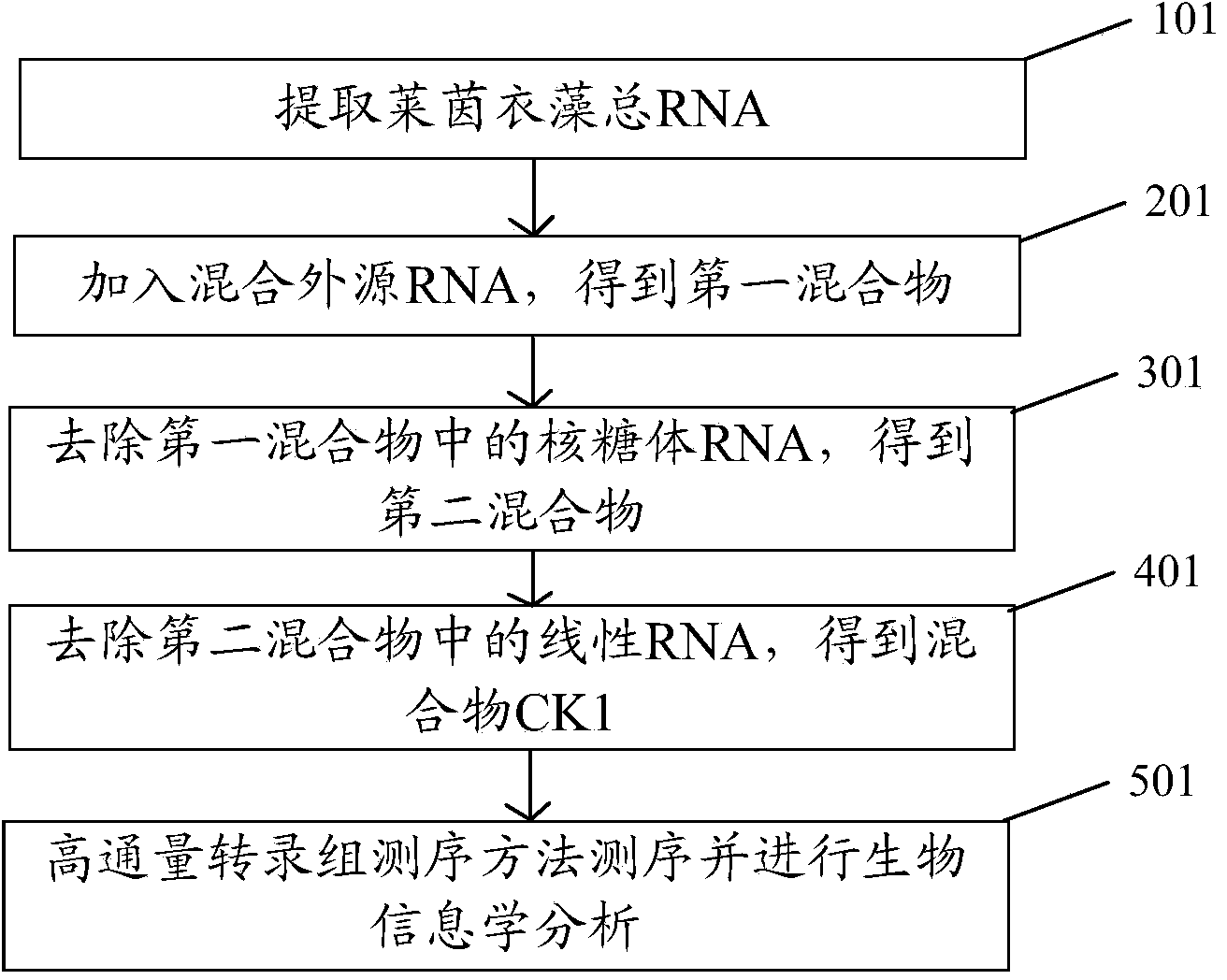
[0038] The embodiment of the present invention provides a new method for circular RNA sequencing. The sample to be sequenced in the embodiment of the present invention is Chlamydomonas reinhardtii, and the strain of Chlamydomonas reinhardtii is CC503 (purchased from the Chlamydomonas Resource Center, the website is : http: / / chlamycollection.org / ), such as figure 1 As shown, the specific methods include:
[0039]Step 100: artificially synthesize exogenous linear ERCC-0004-RNA and exogenous circular ERCC-0013-RNA. Existing circular RNA sequencing uses an exogenous linear RNA as a reference (negative reference). In the embodiment of the present invention, exogenous circular RNA and exogenous linear RNA were artificially synthesized, and total RNA was added in proportion as a reference to simulate endogenous circular RNA and endogenous linear RNA stably expressed in vivo. The exogenous circular RNA and exogenous linear RNA provided by the embodiment of the present invention do n...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com