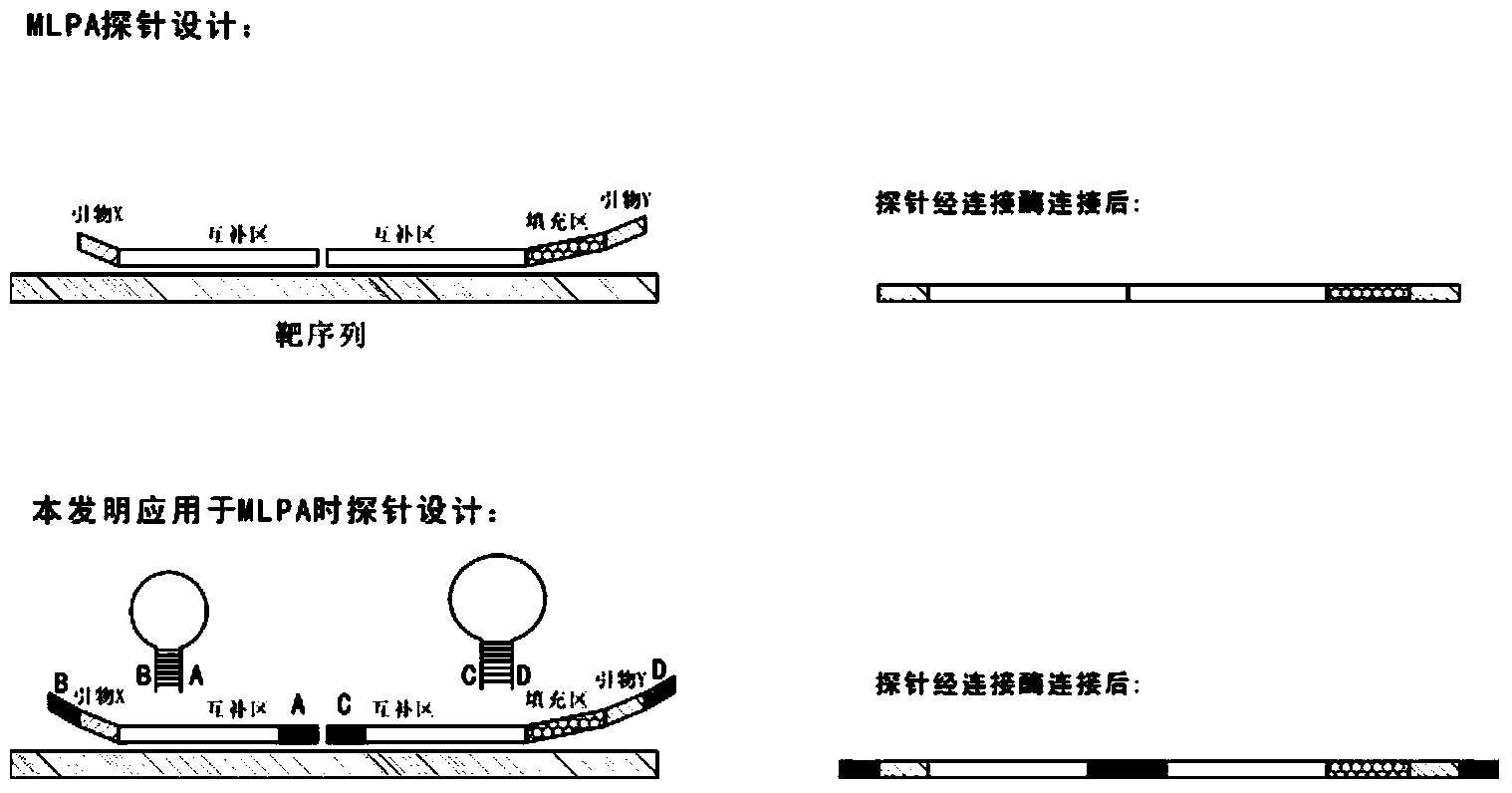
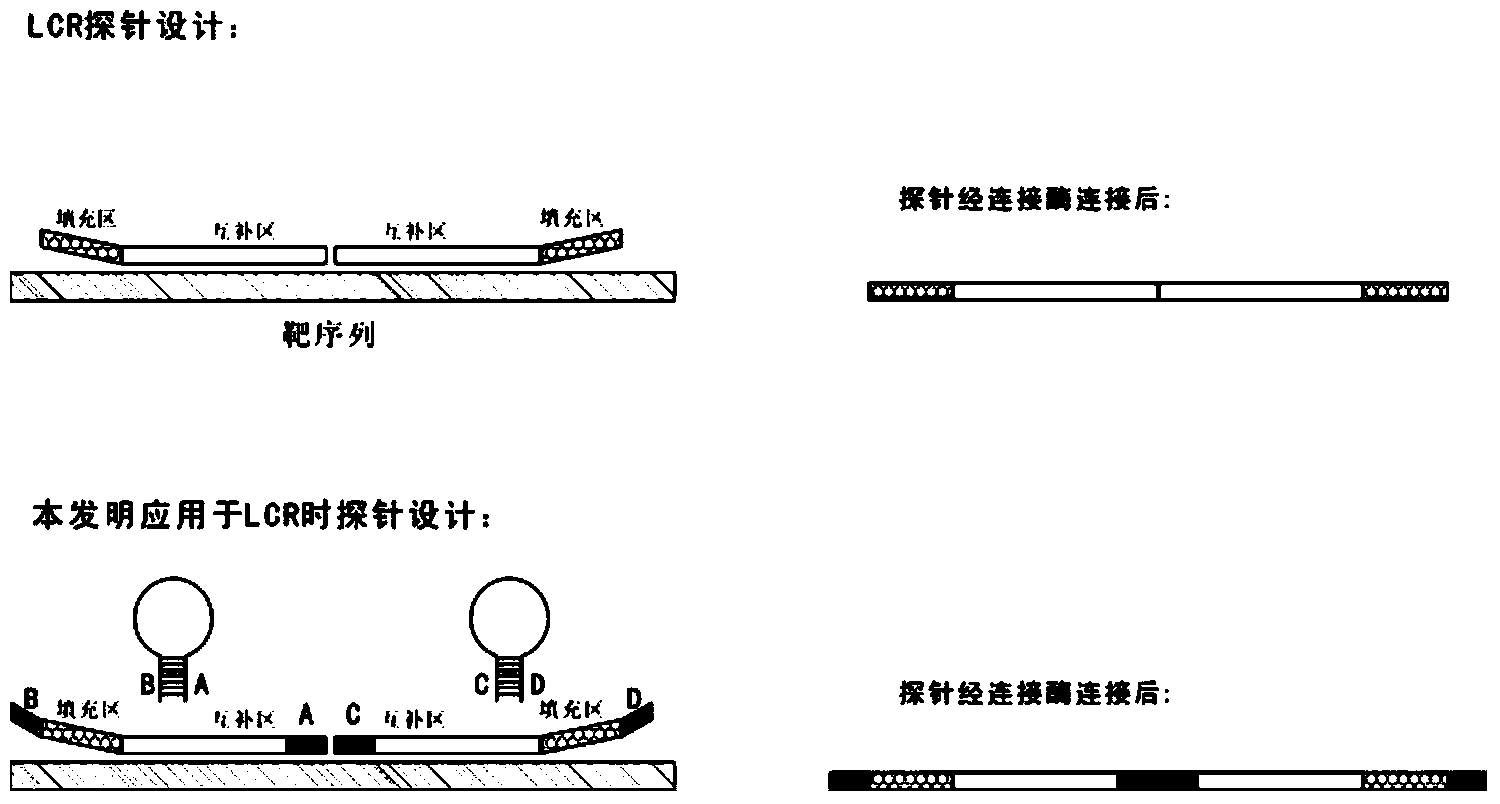
Novel probe design method for ligase reaction
A technology of ligase reaction and design method, which is applied in the field of new probe design for ligase reaction, and can solve problems affecting detection results, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0017] Blood samples from patients with clinically diagnosed non-Hodgkin's lymphoma (NHL) were used as samples. DNA Blood Mini Kit (Qiagen) kit was used to extract blood genomic DNA, and the polymorphic sites rs1800896 and rs1800871 in the promoter region of interleukin 10 were detected by MLPA method.
[0018] According to the probe design method of the present invention, the probe suitable for this detection is designed, and the probe sequence is shown in Table 1:
[0019] Table 1: Probe sequences for detection of rs1800896 and rs1800871
[0020]
[0021] Hybridization: Dissolve the probe in TE to prepare a 5nM solution. Prepare a reaction solution in a PCR tube according to the hybridization reaction system in Table 2, and incubate at 60°C for 16-20 hours for hybridization.
[0022] Ligation: After hybridization, add 3uL of ligase buffer, 0.5uL of ligase, and 6.5uL of ultrapure water into the PCR tube, incubate at 54°C for 20min, heat at 98°C for 5min, and then cool t...
Embodiment 2
[0031] DNA extracted from B lymphocyte LY-01 was used as a sample, and the copy number of gene C1qA was detected by LCR method.
[0032] According to the probe design method of the present invention, the probe suitable for this detection is designed, and the probe sequence is shown in Table 1:
[0033] Table 1: Probe sequences for detecting gene C1qA
[0034]
[0035] LCR reaction: the probe was dissolved in TE to prepare a 5nM solution. Prepare the reaction solution in the PCR tube according to the LCR reaction system in Table 2, place the PCR tube on the PCR instrument, and perform the reaction according to the reaction program in Table 3.
[0036] Table 2: LCR reaction system
[0037]
[0038] Table 3: LCR reaction program
[0039]
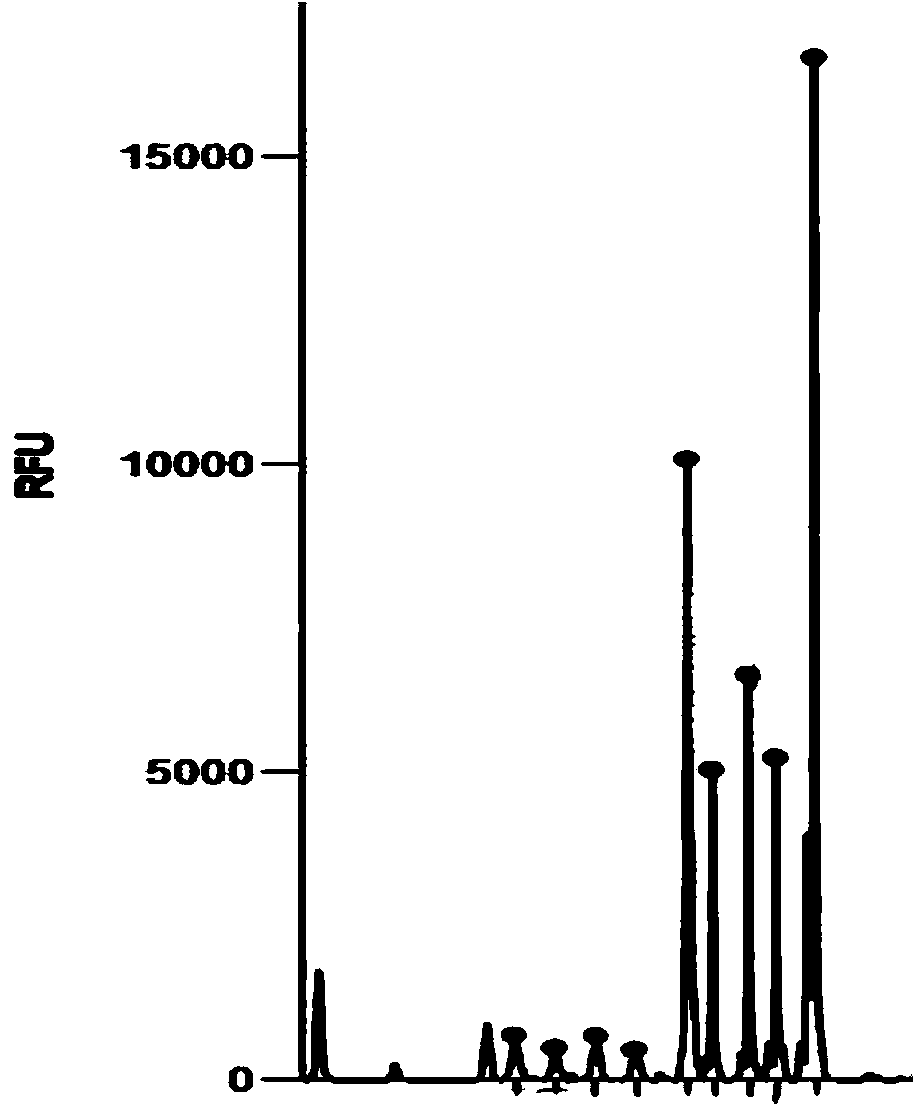
[0040] After the LCR reaction, the amplified product was analyzed by capillary electrophoresis. The length of the amplified fragment of the C1qA gene was 100bp. image 3 . Compared with the copy number of the housekeeping gene, in t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com