A method for detection of complete gene deletion of cyp2a6 based on fluorescent quantitative PCR
A fluorescent quantitative, whole-gene technology, applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., can solve the problems of difficult combination of primers and probes, save experimental time and consumables, ensure accuracy, and improve sensitivity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
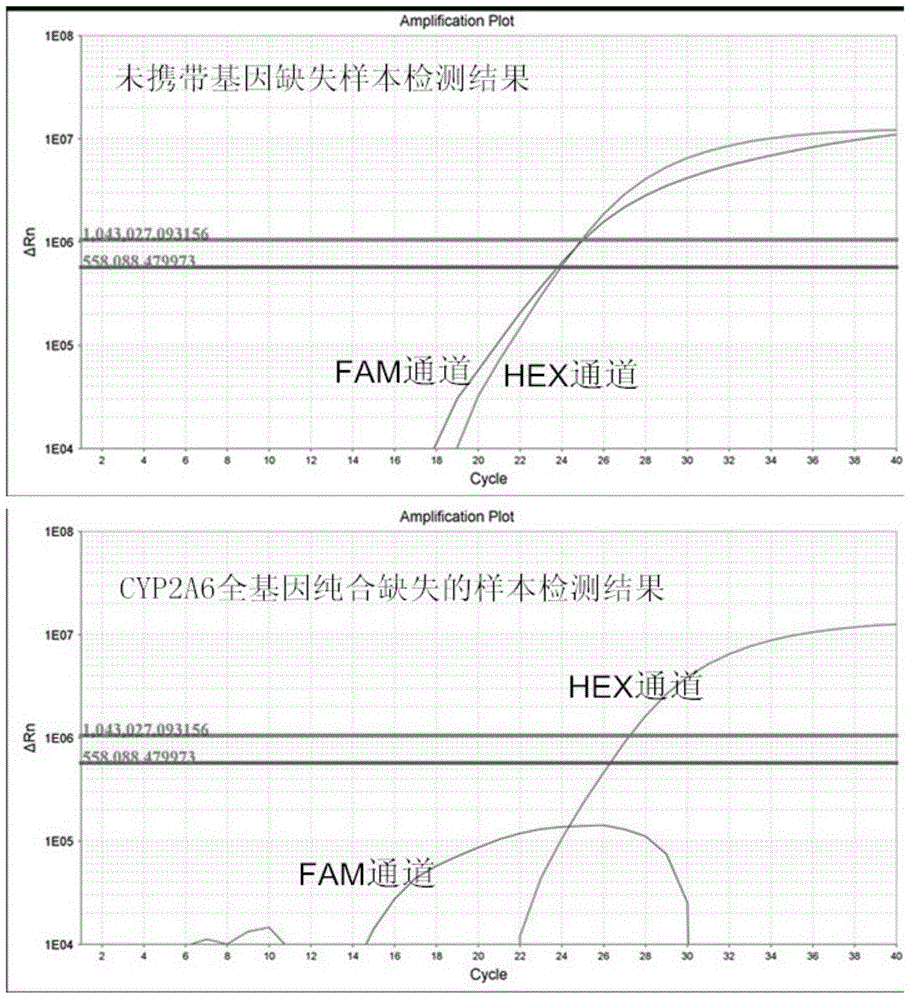
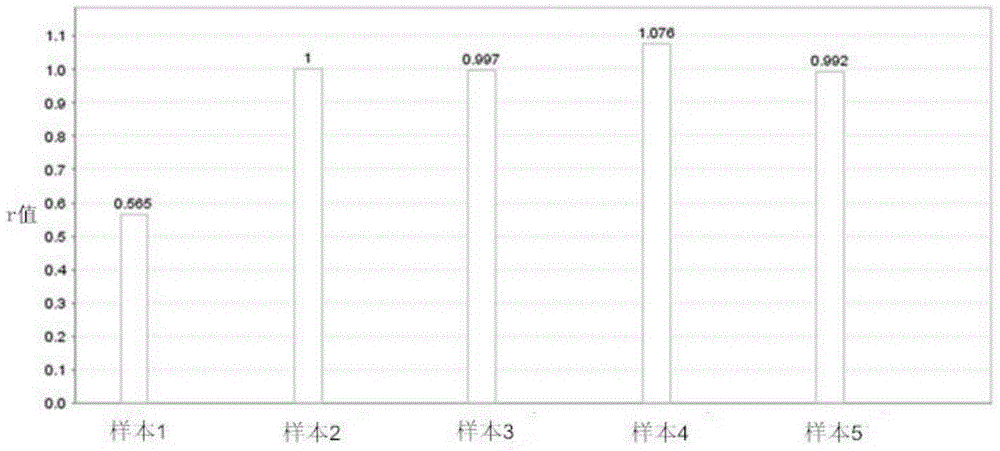
[0042]The present invention provides a method for quickly and accurately detecting CYP2A6 gene deletion based on fluorescent quantitative PCR, that is, by designing highly specific primers for CYP2A6 gene and ALB (as an internal reference gene) respectively, and using fluorescent quantitative PCR technology to quickly and accurately detect CYP2A6 A method for whole gene deletion.
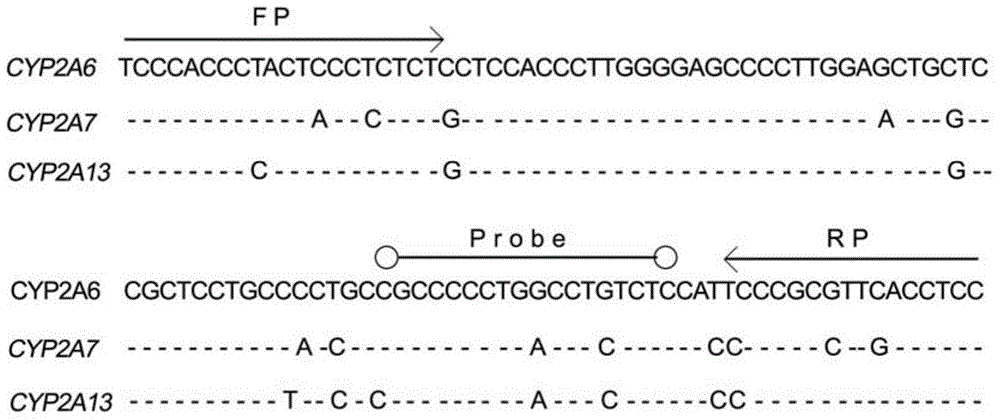
[0043] see figure 1 As shown, the specific primers and probe combinations of the CYP2A6 gene designed in the present invention. Based on the slight difference in the sequences of CYP2A6 gene and CYP2A7 and CYP2A13 genes, and based on the principle that the 3' end sequence of the primer determines its extension efficiency, we designed a set of primer-probe combinations that can amplify the CYP2A6 gene with high specificity. Using this combination for real-time quantitative PCR can not only efficiently amplify the CYP2A6 gene, but also completely avoid the interference of the CYP2A7 and CYP2A13 genes...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com