A method for identifying whether the species to be tested is China Cotton Research Institute 63 and its special primer set
A primer pair and auxiliary identification technology, which is applied in the direction of biochemical equipment and methods, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problems of affecting the promotion effect of hybrid cotton, female parent doping, and unsatisfactory identification results, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Embodiment 1, the construction of the DNA fingerprint collection of China Cotton Research Institute 63
[0030] 1. Screening of SSR primer pairs
[0031] After screening a large number of cotton SSR primers, it was found that 30 SSR primer pairs (see Table 1 for nucleotide sequences) could be used to make the DNA fingerprint of Zhongmian Research Institute 63.
[0032] Nucleotide sequence of table 130 primer pairs
[0033] serial number
Primer name
Forward primer (5'→3')
Reverse primer (5'→3')
1
CIR062
TTTAGAGGAGAAGTTTAGG (sequence 1)
CAGTCTCTTGTAGTTTCATT (sequence 2)
2
CS62
GATGGCTACCTCCCCTTTGTA (sequence 3)
CGTAAGGAAGCCTAGCAAAA (sequence 4)
3
NAU1070
ACCAACAATGGTGACCTCTT (Sequence 5)
CCCTCCATAACCAAAAGTTG (sequence 6)
4
NAU1102
ATCTCTCTGTCTCCCCCCTTC (sequence 7)
GCATATCTGGCGGGTATAAT (sequence 8)
5
NAU1186
AATGGTCCTGCTCCAGATT (sequence 9)
AATCGTCGTCGTCGAAT...
Embodiment 2
[0073] Example 2, application of 30 SSR primer pairs to aid in the identification of China Cotton Research Institute 63
[0074] The following cotton varieties were tested: Zhongzhimian No. 2 (Plant Protection Institute, Chinese Academy of Agricultural Sciences), Ruiza 816 (Jinan Xinrui Seed Industry), Han 8266 (Handan Academy of Agricultural Sciences), Zhongmian Institute 63 ( China Cotton Seed Industry).
[0075] Detection method: extract the genomic DNA of cotton leaves, use the genomic DNA as a template, carry out PCR amplification with the 30 SSR primer pairs respectively, and then perform 8% polyacrylamide gel electrophoresis and silver staining for color development.
[0076] The PCR system and PCR reaction procedures are the same as in Example 1.
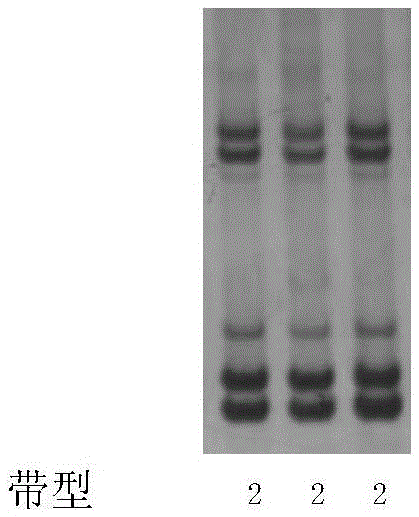
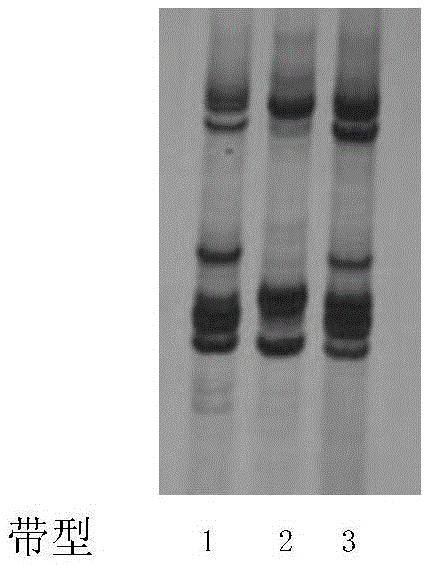
[0077] Electropherogram see Figure 31 (1, 2, 3, and 4 represent Zhongmian Institute 63, Zhongzhimian No. 2, Ruiza 816, and Han 8266, respectively). The band results are shown in Table 3.
[0078] Table 3 adopts 30 pairs...
Embodiment 3
[0113] Embodiment 3, application 20 primer pairs in 30 SSR primer pairs identify seed purity
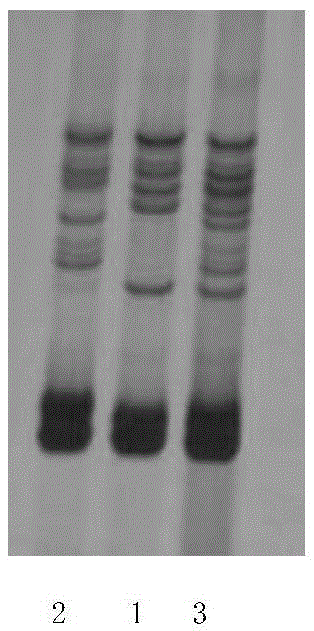
[0114] Receive a batch of China Cotton Institute 63 (in the form of seeds, a total of 200 grains from the seed production company; the seeds are theoretically China Cotton Institute 63, but usually due to the limited operating level in the seed production process, it is difficult to achieve 100% purity. female parent plant phenomenon or other doping phenomenon), randomly select 26 seeds from 200 seeds, extract the genomic DNA of the kernel respectively, use the genomic DNA as a template, and use 20 SSR primer pairs respectively (see the table for specific SSR primer pairs) 4) Perform PCR amplification, and then perform 8% polyacrylamide gel electrophoresis and silver staining for color development. Using the genomic DNA of the male parent (2 repeated samples) of Zhongmian Institute 63 as a template, 20 SSR primer pairs were used for PCR amplification, and then 8% polyacrylamide gel e...
PUM
| Property | Measurement | Unit |
|---|---|---|
| elongation at break | aaaaa | aaaaa |
| reflectance | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com