Primer set, kit and method for detecting maize mir162 by lamp
A detection method and primer set technology, applied in the field of molecular biology, can solve the problems of detection methods and kits for undiscovered transgenic maize MIR162 strains, achieve broad application value and market prospects, simple result determination, and short detection time Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Embodiment 1: the detection of corn sample
[0044] 1. Primer design and synthesis:
[0045] According to the gene sequence of the transgenic maize line MIR162 (Genbank sequence number HI203349), the LAMP primer set was designed. The LAMP primer set consisted of outer primer F3, inner primer FIP, inner primer BIP, outer primer B3, loop primer LF, and loop primer LB. The specific sequence is as follows:
[0046] Outer primer F3: 5'-ACATGTAATGCATGACGTTAT-3'SEQ ID No: 1
[0047] Outer primer B3: 5'-GCCTTATCTGTTGCCTTCA-3'SEQ ID No: 2
[0048] Inner primer FIP:
[0049] 5'-CGCTATATTTTGTTTTTCTATCGCGTGAGATGGGTTTTTTATGATTAGAGTC-3'
[0050] SEQ ID No: 3
[0051] Inner primer BIP:
[0052] 5'-AACTAGGATAAATTATCGCGCGCCGGACGTTTTTAATGTACTGAA-3'SEQ ID No: 4
[0053] Loop primer LB: 5'-TGTCATCTATGTTACTAGATCCCCG-3'SEQ ID No: 5
[0054] The above primers were synthesized by TAKARA Company. The outer primer F3 solution, the outer primer B3 solution of 10 μmol / L, the inner pri...
Embodiment 2
[0066] Example 2: Determination of the specificity of the LAMP method for the detection of transgenic maize MIR162 lines
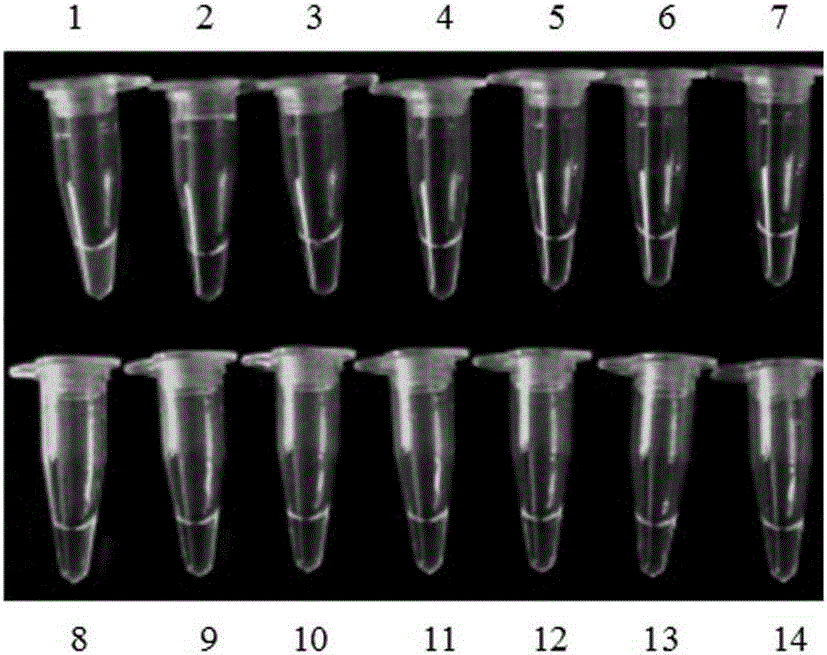
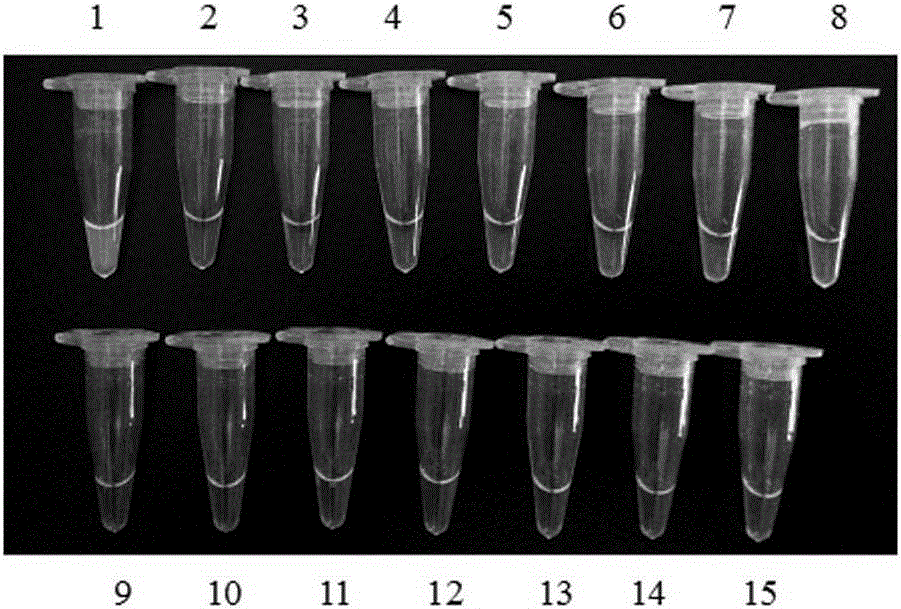
[0067] Take 13 parts of seeds or fruits of other plant species, 14 parts of standard samples of different transgenic maize lines, and 1 part of non-transgenic maize. Genomic DNA was extracted according to the above method, and subjected to LAMP isothermal amplification, and the fluorescent dye SYBRGreenI colorimetric method was used to detect the results. The results showed that only the transgenic maize MIR162 line produced green specifically, and the test results were as follows: figure 1 and figure 2 shown.
[0068] See Example 1 for the primer sets and methods used.
[0069] figure 1 It is a diagram of the experimental results of detecting MIR162 and other 13 plant species by using the primer set and detection method of the present invention; Tubes 1-14 are sequentially transgenic corn MIR162 (10%, that is, the ratio of the mass of transgenic c...
Embodiment 3
[0072] Example 3: Determination of the sensitivity of the LAMP method for the detection of transgenic maize MIR162 lines
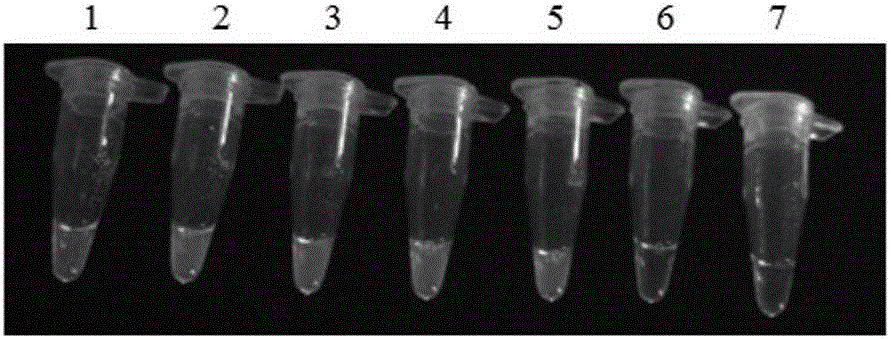
[0073] The transgenic corn line MIR162 standard was mixed with non-transgenic corn seeds to prepare the relative percentage (w / w) of the transgenic corn line MIR162 as 100%, 10%, 5%, 1%, 0.5%, 0.1%, 0% Genomic DNA was extracted respectively according to the methods of the above-mentioned examples, and then subjected to isothermal amplification, and the results were detected by chromogenic method. Test results such as image 3 shown. image 3 It is a diagram of the sensitivity test results of the LAMP detection method of the transgenic maize MIR162 strain of the present invention. Tubes 1 to 7 are the LAMP test results of the corn seed samples with transgenic corn line MIR162 content respectively 100%, 10%, 5%, 1%, 0.5%, 0.1%, and 0%.
[0074] See Example 1 for the primer sets and methods used.
[0075] The experimental results showed that the samples wit...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com