Processes and compositions for methylation-based enrichment of fetal nucleic acid from a maternal sample useful for non invasive prenatal diagnoses
A methylation and nucleic acid technology, applied in biochemical equipment and methods, disease diagnosis, biomaterial analysis, etc., can solve problems such as maternal and child risks
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
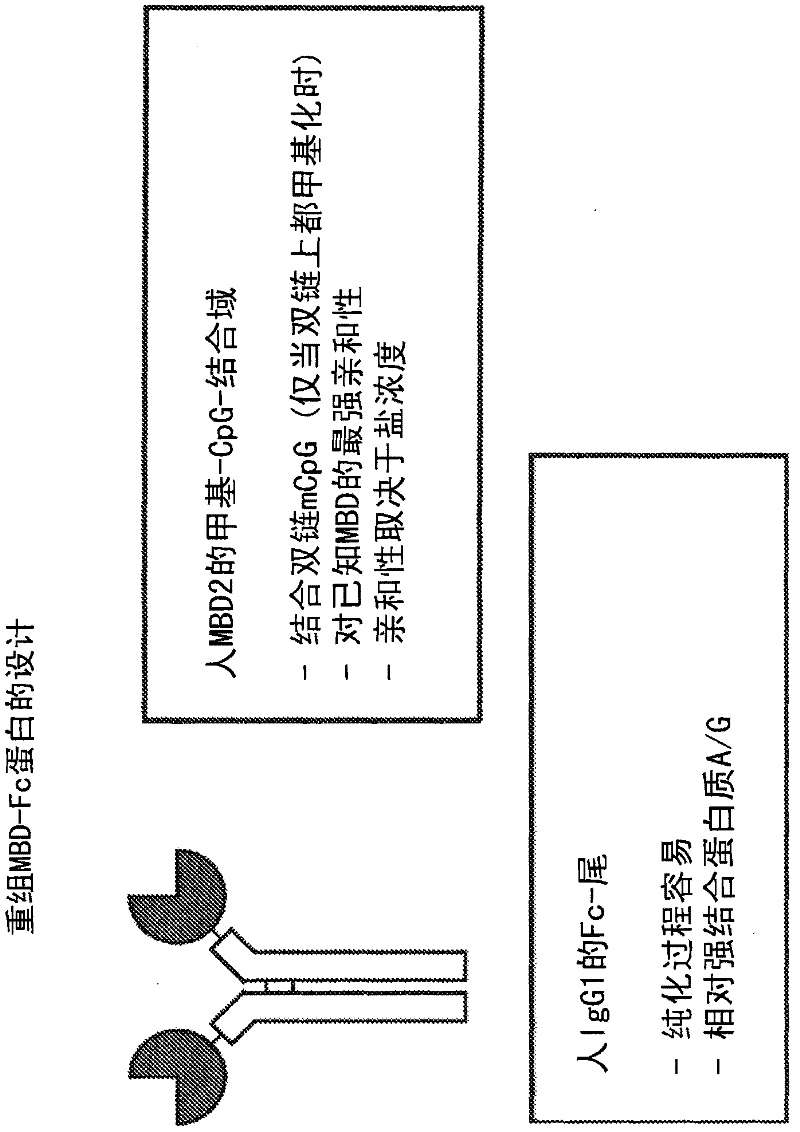
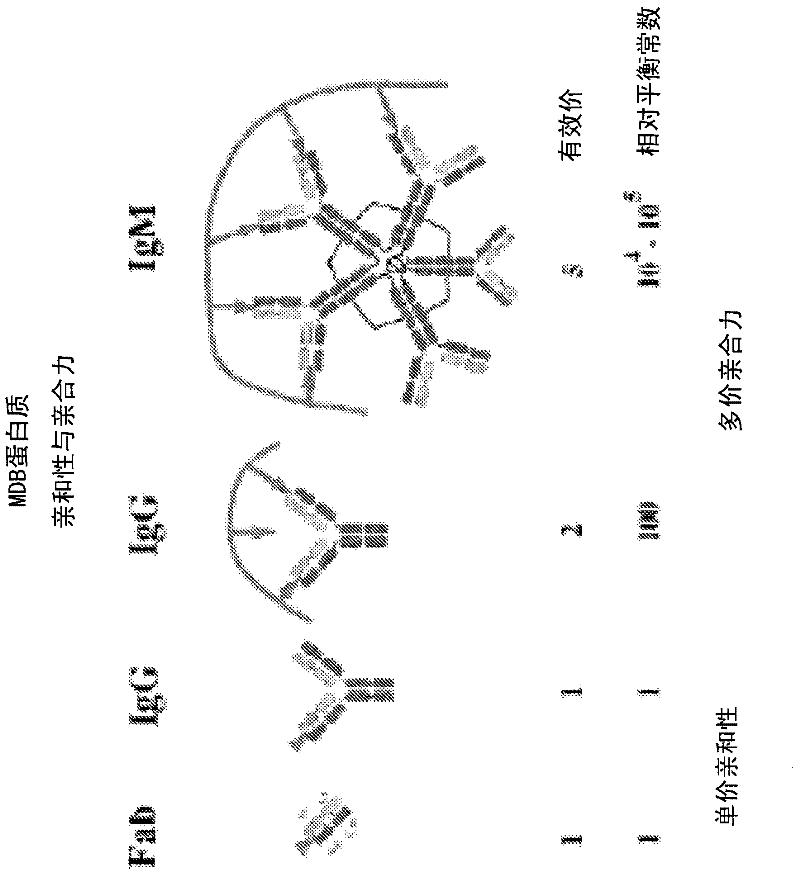
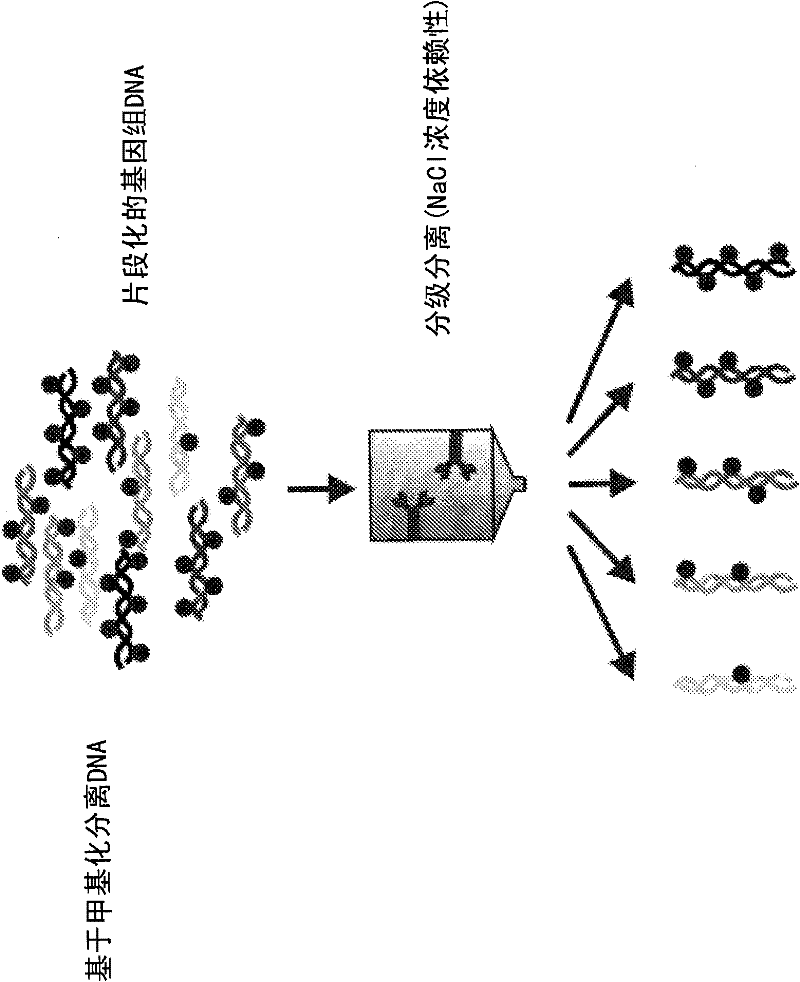
[0197] In the following examples, 10 paired maternal and placental DNA samples were used to identify differentially methylated regions. These results were confirmed using a mass spectrometry-based quantitative methylation assay. First, genomic DNA was extracted from maternal buffy coat and corresponding placental tissue. Next, the methylated fraction of each DNA sample was captured with MBD-FC. See Figure 1-3 . The two tissue sections were labeled with different fluorescent dyes and compared with the Agilent CpG island microarray hybridization. See Figure 4 . This is done by identifying differentially methylated regions that can be used for prenatal diagnosis. Therefore, two criteria were used to select genomic regions as potential enrichment markers: the observed difference in methylation must be present in all tested sample pairs, and the length of the region must be greater than 200 bp.
[0198] DNA preparation and fragmentation
[0199] Genomic DNA (gDNA) from ...
Embodiment 2
[0219] Example 2 describes a non-invasive protocol for detecting the amount of fetal nucleic acid present in a maternal sample (referred to herein as a "fetal quantification method") that can be used to detect or confirm a fetal trait (e.g., fetal sex for RhD compatibility) or Diagnosis of chromosomal abnormalities such as trisomy 21 (both are referred to herein as "methods of methylation-based fetal diagnosis"). Figure 10 shows an embodiment of the fetal quantification method, while Figure 11 One embodiment of a method for methylation-based fetal diagnosis is shown. Both methods use fetal DNA obtained from a maternal sample. Samples contain differentially methylated maternal and fetal nucleic acids. For example, the sample can be maternal plasma or serum. Fetal DNA accounts for approximately 2-30% of the total DNA in maternal plasma. The exact amount of fetal contribution to the total nucleic acid present in a sample varies from pregnancy to pregnancy and can vary based...
Embodiment 3
[0323] Example 3 - Other differentially methylated targets
[0324] Differentially methylated targets not located on chromosome 21
[0325] Additional differentially methylated targets were selected for further analysis based on previous microarray analysis. See Example 1 for a description of the microarray analysis. In microarray screening, differentially methylated regions (DMRs) between placental tissue and PBMCs were defined. Regions were selected for EpiTYPER confirmation based on hypermethylation in placenta relative to PBMC. After selecting the orientation of the change, regions are selected based on statistical significance, with the design regions starting at the highest significance and progressing downward according to significance. These studies were performed in 8 paired PBMC and placenta samples. Additional non-chromosomal 21 targets are provided in Table IB, along with representative genomic sequences for each target in Table 4B.
[0326] Differentially met...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com