Method and device for predicting three-dimensional protein structure
A prediction method and a technology of three-dimensional structure, applied in special data processing applications, instruments, electrical digital data processing, etc., can solve the problem of low accuracy, achieve the effect of improving accuracy, good prediction effect, and reducing preference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
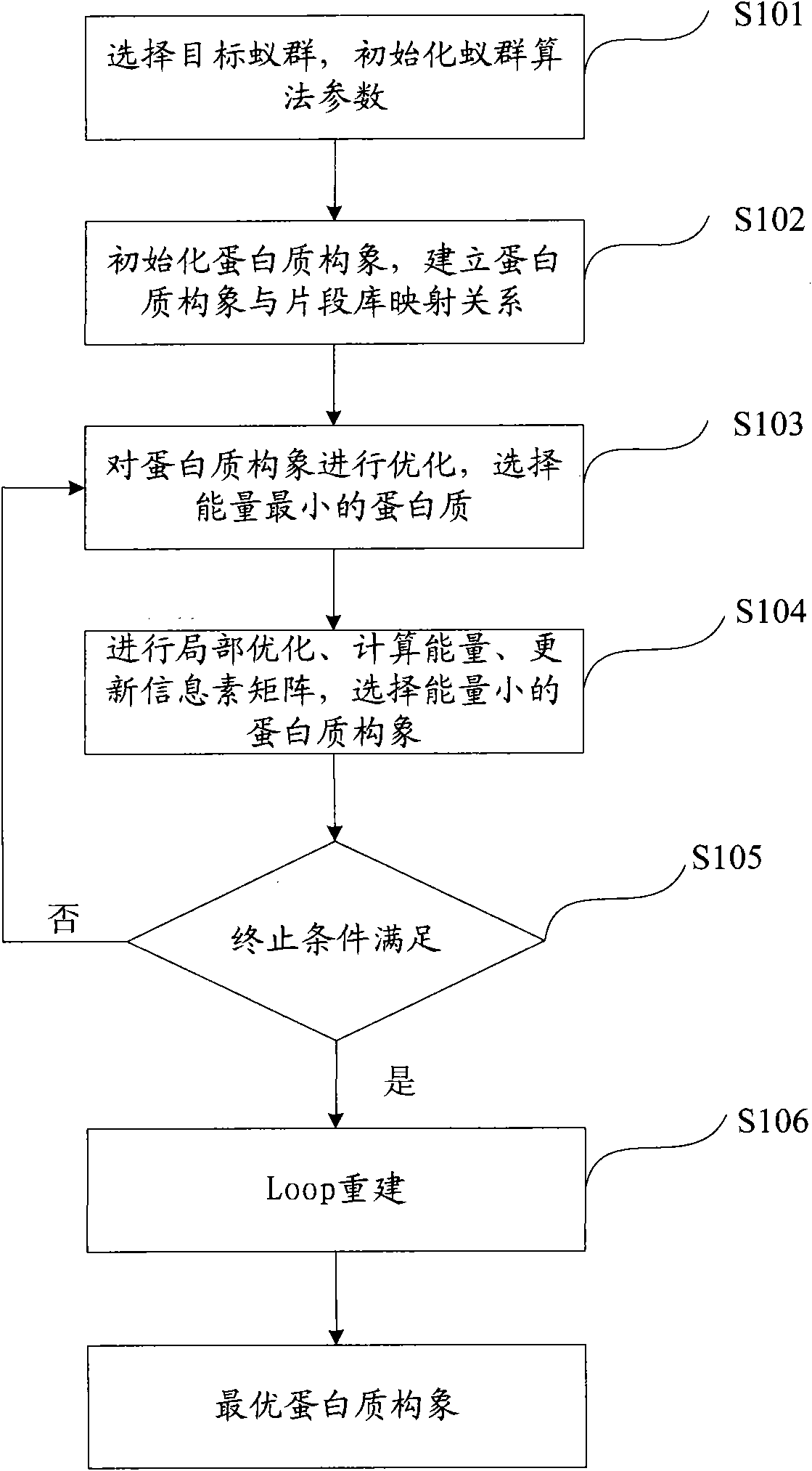
[0059] see figure 1 , figure 1 It is a flow chart of the method for predicting the three-dimensional structure of a protein provided in Example 1 of the present invention.
[0060] Step S101, select the target ant colony, and initialize the parameters of the ant colony algorithm.
[0061] Step S101 is executed to select the target ant colony, that is, to determine the number of the target ant colony. In this embodiment, the number of ant colonies is determined to be 1, that is, the prediction based on the single ant colony algorithm is performed.
[0062] Initialize the ant colony algorithm parameters, the ant colony algorithm parameters include:
[0063] probability parameter q 1 ; In the present invention, the probability parameter q 1 The value range of is [0, 1], which is set to 0.8 in this embodiment;
[0064] Pheromone τ, its initial value is set to 10 in this embodiment;
[0065] The pheromone volatilization coefficient ρ, in the present invention, the value range...
Embodiment 2
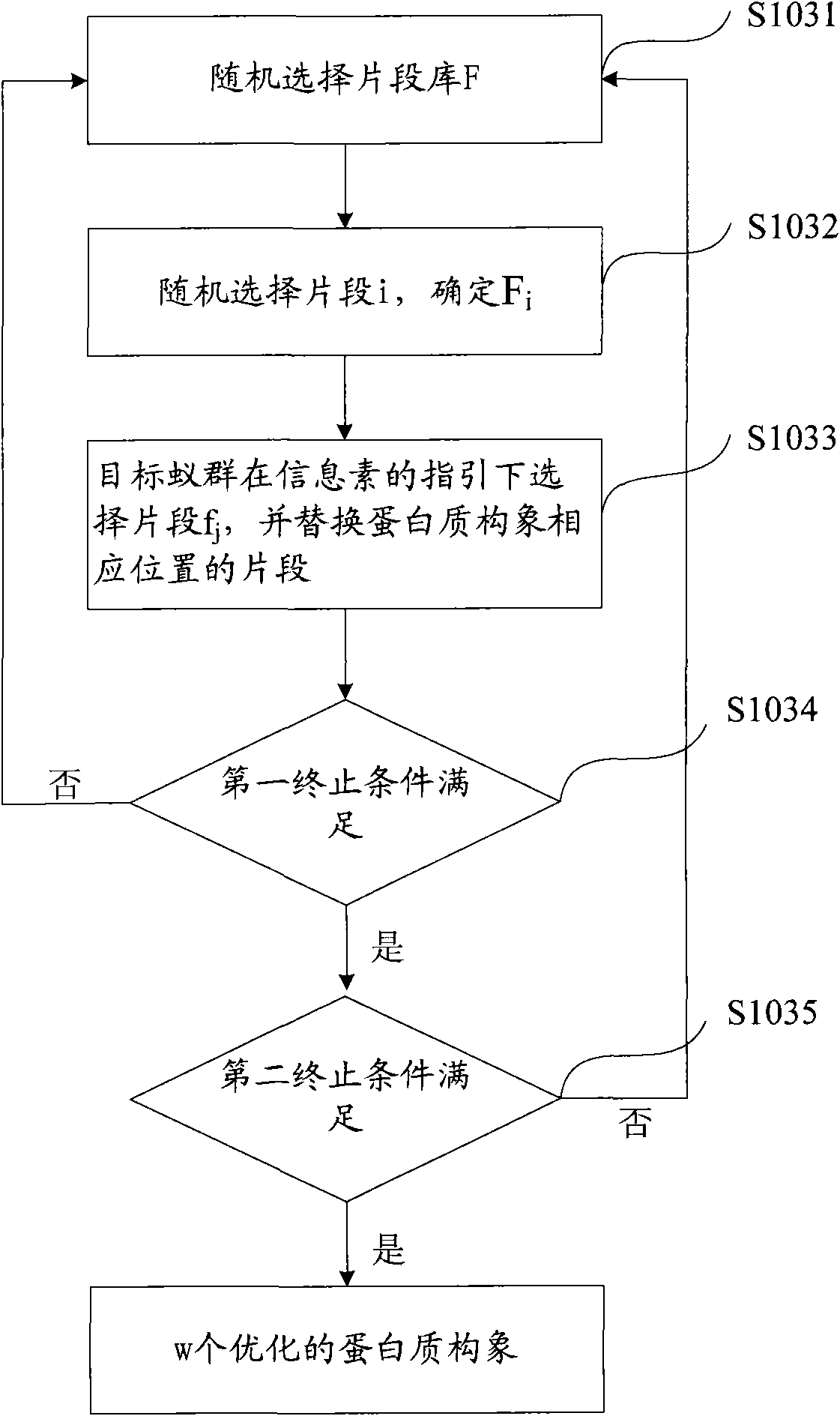
[0149] On the basis of the disclosure of the first embodiment, the present invention also discloses an embodiment, see Figure 4 , Figure 4 It is a flowchart of the method for predicting the three-dimensional structure of a protein provided in Example 2 of the present invention.
[0150] Different from the first example, in the second example, when selecting the target ant colony, p ant colonies are selected, and p>1, that is, in the second example, multiple ant colonies are used to predict the protein structure in parallel. When selecting the target ant colony, the number of the target ant colony can be determined according to the number of processors. In this embodiment, the number of ant colonies is determined to be 8.
[0151] When initializing the parameters of each ant colony algorithm, the 8 target ant colonies share the same pheromone matrix, that is, the 8 target ant colonies share the same pheromone matrix to predict the three-dimensional structure of the same prot...
Embodiment 3
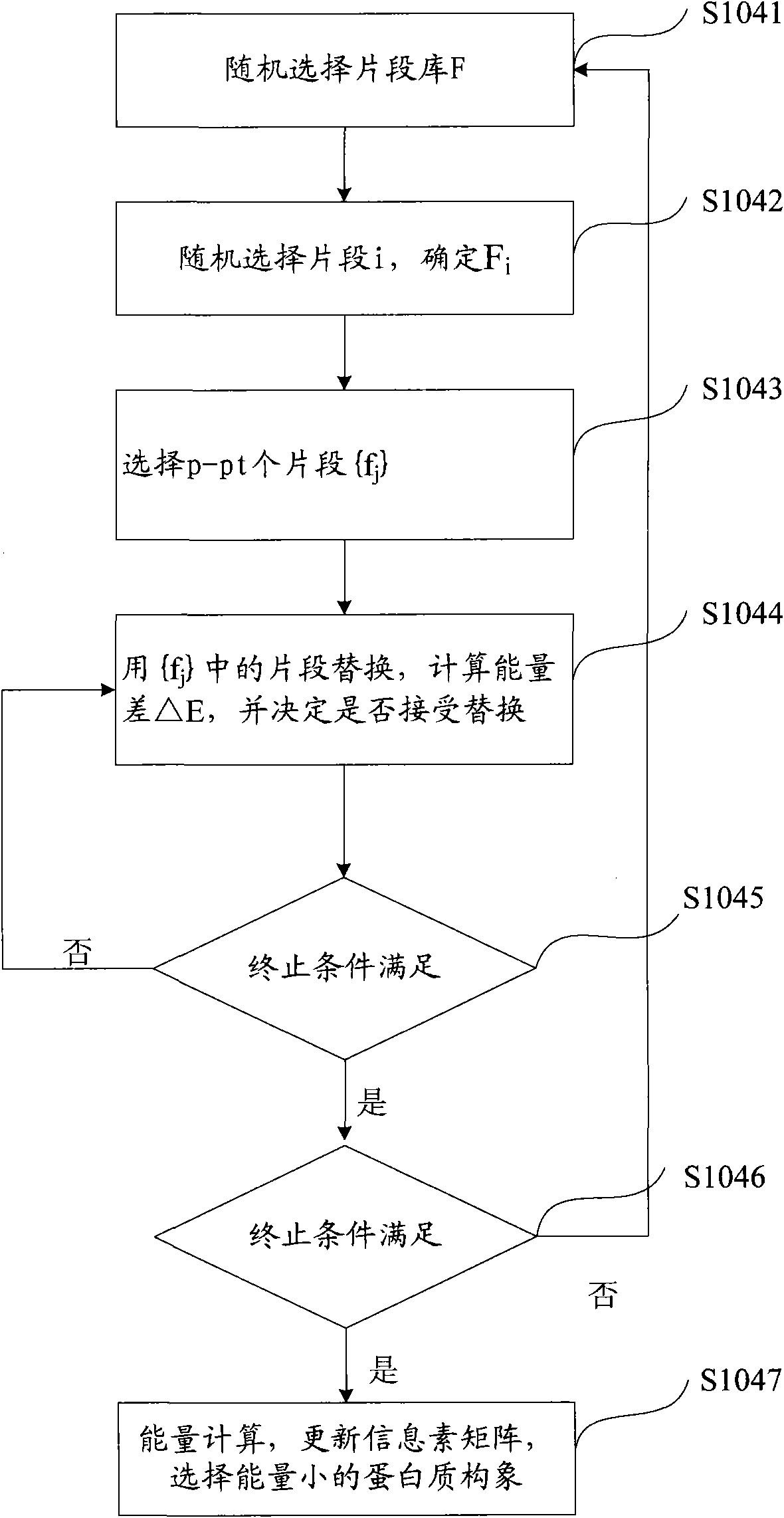
[0157] On the basis of the disclosure of the second embodiment, the present invention also discloses an embodiment, see Figure 5 , Figure 5 It is a flowchart of the method for predicting the three-dimensional structure of a protein provided in Example 3 of the present invention.
[0158] After using the parallel ant colony prediction method to obtain p optimal protein conformations, step S107 is also included: performing crossover on the p protein conformations to quickly obtain p crossover protein conformations. The crossover method is specifically:
[0159] Step S1071, randomly select two protein conformations M among the p optimal protein conformations bi and M bj is the parent conformation;
[0160] Step S1072, randomly in the parent conformation M bi and M bj Determine the position k, take the position k as the cut-off point, and set the parent conformation M bi cut into M bi k- and M bi K+ , the parent conformation M bj cut into M bj k- and M bj K+ ;
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com