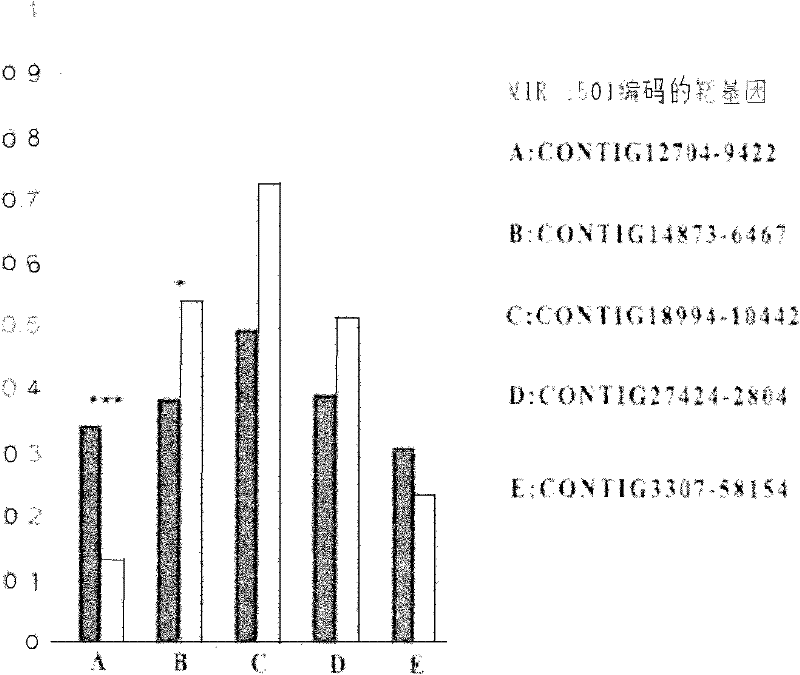
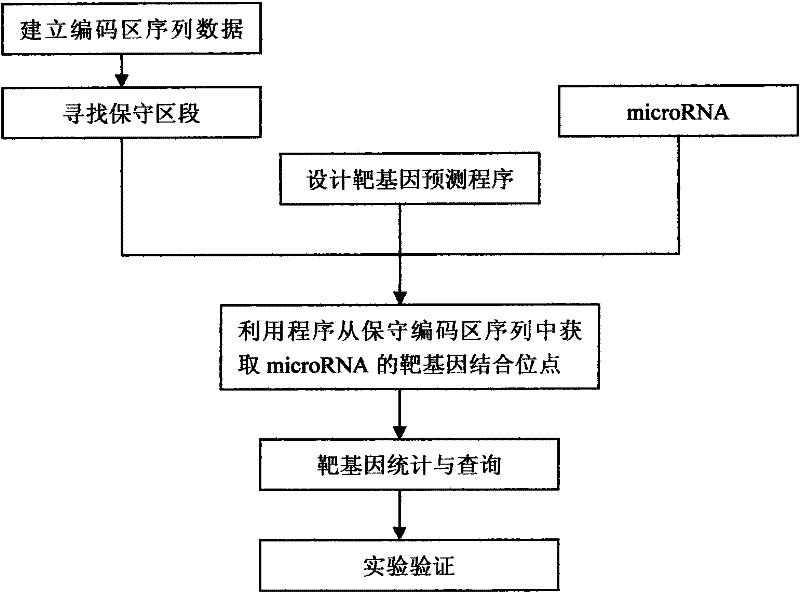
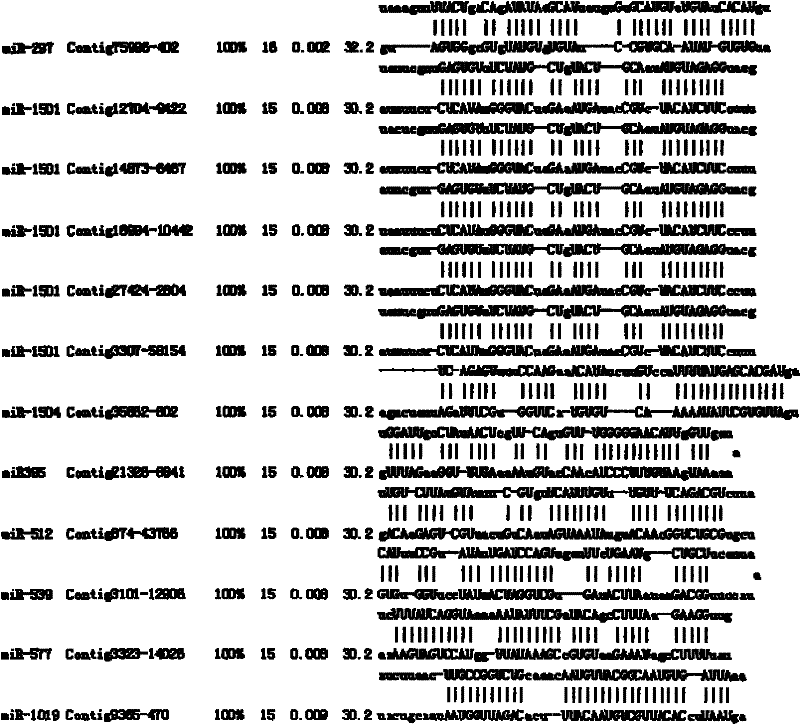
A method for target gene prediction in microRNA coding region
A coding region and target gene technology, applied in special data processing applications, instruments, electrical digital data processing, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0025] We take the target gene prediction of the planarian coding region as an example to illustrate the implementation process of the method of the present invention:
[0026] (1) Establish a coding region sequence database
[0027] The mRNA sequence of Schmidteamediterranea was downloaded from http: / / www.ncbi.nlm.nih.gov. By writing a PERL script program to obtain the coding sequence of the protein from the mRNA sequence, a coding region sequence database was established.
[0028] (2) Align the coding region sequences in mammals to find conserved segments.
[0029] Using the online BLAST program (http: / / www.ncbi.nlm.nih.gov / blast) to compare the sequences of mammalian coding regions, remove the poorly conserved coding region sequences, and proceed to the next step.
[0030] (3) Obtain the sequence information of planarian microRNA
[0031] Download all known mature microRNA sequences of planarians from http: / / microrna.sanger.ac.uk / sequences /
[0032] (4) Prediction of ta...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com