Cryophilic xylosidase/arabinofuranosidase and preparation method and application thereof
A technology of arabinosidase and xylosidase, applied in the field of cold-adapted xylosidase/arabinosidase bifunctional cellulose degrading enzyme and its preparation, to achieve the effects of improving utilization rate, mild conditions and increasing yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Construction of yak rumen microbial metagenomic cosmid library and screening of xylosidase activity
[0047] The rumen samples of yaks in Qinghai, China were collected, filtered with 3 layers of gauze, and the filtrate was centrifuged to collect rumen microbial cells. For the extraction method of rumen microbial metagenome, please refer to An et al. ("Anaerobe" 2005, Volume 11, pages 207-215). For the construction method of the rumen-uncultured bacterial metagenomic library, refer to the product manual of the pWEB::TNC Cosmid Cloning Kit kit from Epicentre Company. The extracted metagenomic DNA was blunted with End-Repair Enzyme Mix, ligated with the dephosphorylated vector pWEB::TNC in the kit, and the ligated product was used MaxPlax TM After the Lambda Packaging Extracts were packaged, the host bacteria E.coli EPI100 was infected, and then spread on the LB-ampicillin plate, and cultured at 37°C for 12-16 hours to grow colonies, which were the metagenomic library of ...
Embodiment 2
[0048] Cloning and analysis of the sequence of the RuBGX1 gene of embodiment 2 rumen microorganism source
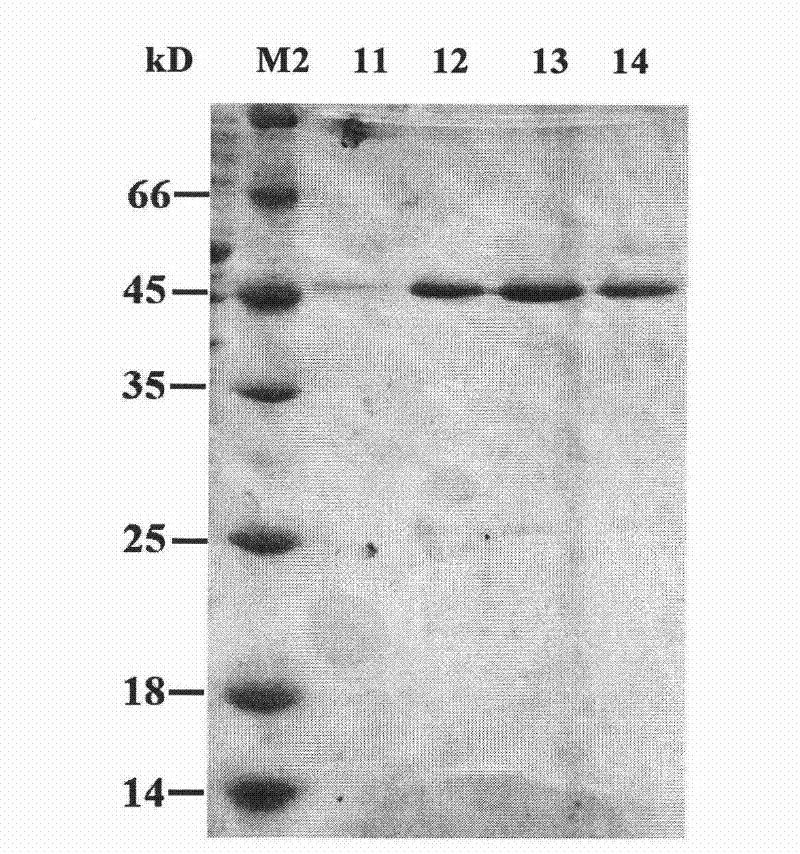
[0049] The screened cosmid-positive library clones were subcloned in pGEM11z, functionally screened, and sequenced. Specifically: the cosmid plasmid of the screened positive clone was partially digested with Sau3AI into a 2-5kb fragment, ligated into the pGEM11z vector that was digested with BamHI and dephosphorylated, and transformed into DH5α. The subcloned library was screened for function, and the obtained subclones were sequenced with T7 and SP6 universal primers, and the sequence of the coding region of the β-1,4-xylosidase gene was analyzed by the analysis software DNAMAN, and the nucleotide sequence of the gene was determined, such as SEQ ID NO 1 and named RuXyn1. RuXyn1 encodes 332 amino acids, its amino acid sequence is shown in SEQ ID NO.2, and its theoretical molecular weight is 42kDa. The protein domain of RuXyn1 was analyzed using SMART software (http: / / w...
Embodiment 3
[0051] Recombinant Expression of RuXyn1 Gene in Escherichia coli
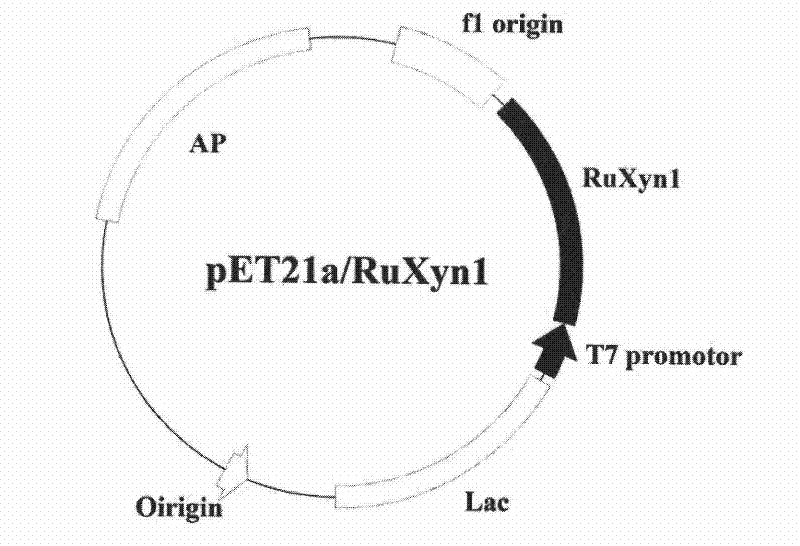
[0052] The recombinant expression of RuXyn1 in Escherichia coli adopts the 6His fusion expression strategy, and the Escherichia coli expression vector pET-21a(+) (purchased from Novagen Company) is selected. Specifically: design and synthesize a set of oligonucleotide primers: forward primer 21axyn1FE: GGC GAATTC and reverse primer 21axyn1FX:CCG CTCGAG (The underlined parts in the sequence are respectively the restriction endonuclease EcoRI and the restriction endonuclease EcoRI and the restriction site of XhoI, and gene sequence is expressed in italics), by PCR amplifying RuXyn1 gene, after recovering by agarose electrophoresis, digest with EcoRI and XhoI respectively , connected with the vector pET-21a(+) digested with EcoRI and XhoI, the schematic diagram of the constructed RuXyn1 recombinant expression vector is as follows figure 1 shown. The ligation product was transformed into Escherichia coli T...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com