Genes frequently altered in pancreatic neuroendocrine tumors
a neuroendocrine tumor and pancreatic gene technology, applied in the field of pancreatic neuroendocrine tumors, can solve the problems of insufficient information about this tumor and relatively ineffective medical therapies
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
lection, Preparation, and Decoding
[0032]To gain insights into the genetic basis of this tumor type, we determined the exomic sequence of 18,000 protein-coding genes in a Discovery set of ten well-characterized sporadic PanNETs. A clinically homogeneous set of tumors of high neoplastic cellularity is essential for the successful identification of genes and pathways involved in any tumor type. Thus, we excluded small cell and large neuroendocrine carcinomas and studied only samples that were not part of a familial syndrome. We macrodisected them to achieve a neoplastic cellularity of >80%. DNA from the enriched neoplastic samples and from matched non-neoplastic tissue from ten patients was used to prepare fragment libraries suitable for massively parallel sequencing. The coding sequences were enriched by capture with the SureSelect Enrichment System and sequenced using an Illumina GAIIx platform (10). The average coverage of each base in the targeted regions was 101-fold and 94.8% of ...
example 2
Analysis in Discovery Set
[0033]We identified 157 somatic mutations in 158 genes among the ten tumors used in the Discovery set. The mutations per tumor ranged from 8 to 23, with a mean of 16 (table S2). There were some obvious differences between the genetic landscapes of PanNETs and those of pancreatic ductal adenocarcinomas (PDAC, ref 11). First, there were 60% fewer genes mutated per tumor in PanNETs than in PDACs. Second, the genes most commonly affected by mutation in PDACs (KRAS, TGF-β pathway, CDKN2A, TP53) were rarely altered in PanNETs and vice versa (table S3). Third, the spectrum of mutations in PDAC and PanNET were different, with C to T transitions more common in PDACs than in PanNETs, and C to G transversions more common in PanNETs than in PDACs (table S4). This suggests that PanNETs are exposed to different environmental carcinogens or that they harbor different repair pathways than PDACs.
example 3
Analysis in Validation Set
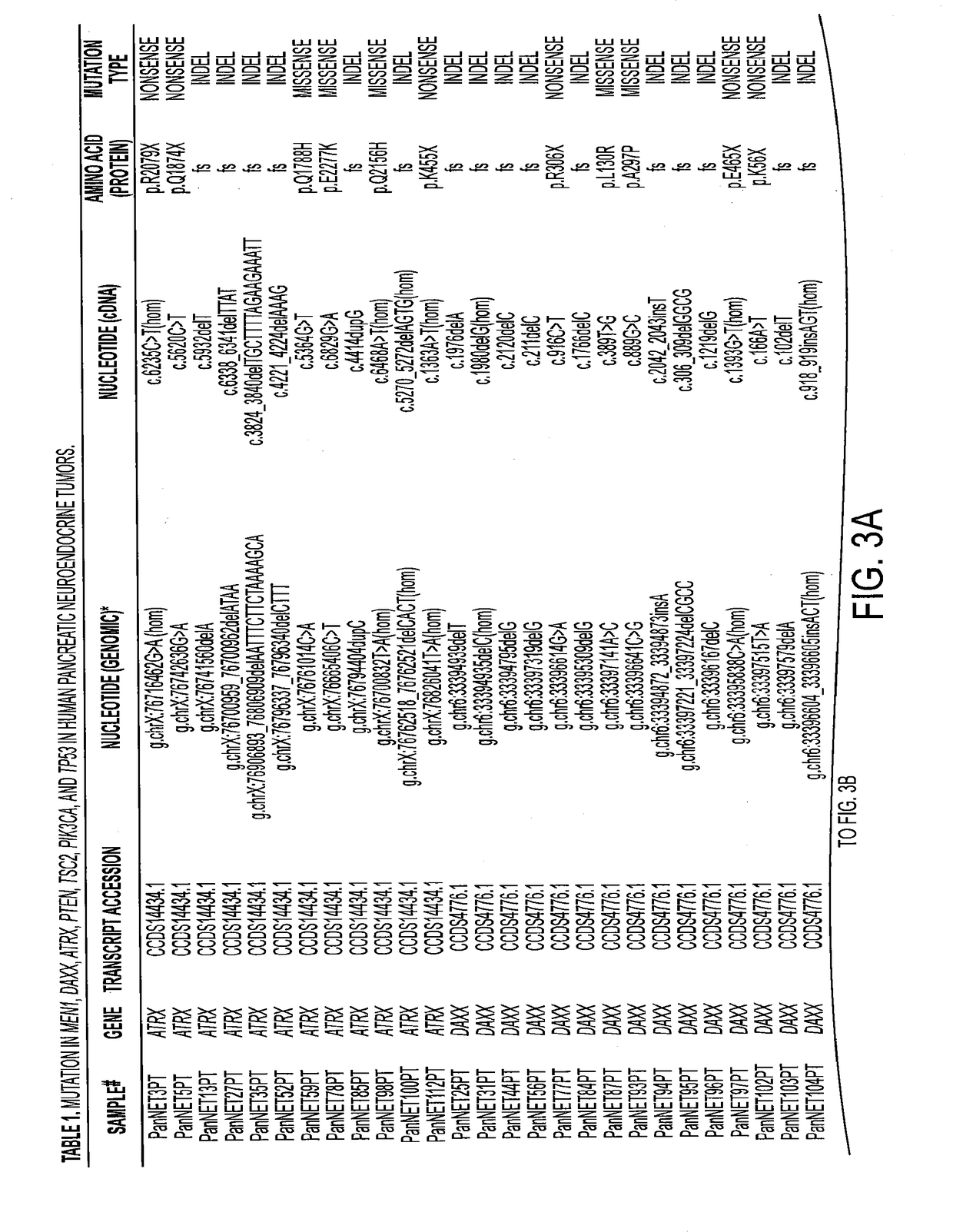
[0034]Four genes were mutated in at least two tumors in the Discovery set: MEN1 in five, DAXX in three, PTEN in two, and TSC2 in two. Somatic mutations in each of these genes were confirmed by Sanger sequencing. The sequences of these genes were then determined by Sanger sequencing in a Validation set consisting of 58 additional PanNETs and their corresponding normal tissues (FIG. 1a,b). Although ATRX was mutated in only one sample in the Discovery set, it was included in the list of genes for further evaluation in the Validation set because its product forms a heterodimer with DAXX and therefore is part of the same pathway. Similarly, PIK3CA was included because it is considered to be part of the mTOR pathway that includes PTEN and TSC2 (12-14). In total, somatic mutations in MEN1, DAXX, ATRX, PTEN, TSC2, and PIK3CA were identified in 44.1%, 25%, 17.6%, 7.3%, 8.8%, and 1.4% PanNETs, respectively (Table 1).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com