Targeting sequence for paenibacillus-based endospore display platform
A Bacillus genus, sequence technology, applied to methods using spores, methods based on microorganisms, immobilized on or in biological cells, etc., can solve the problems that cannot be directly converted to the Bacillus family
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0246] Example 1: General protocol for identifying collagen-like spore surface proteins suitable for endospore display
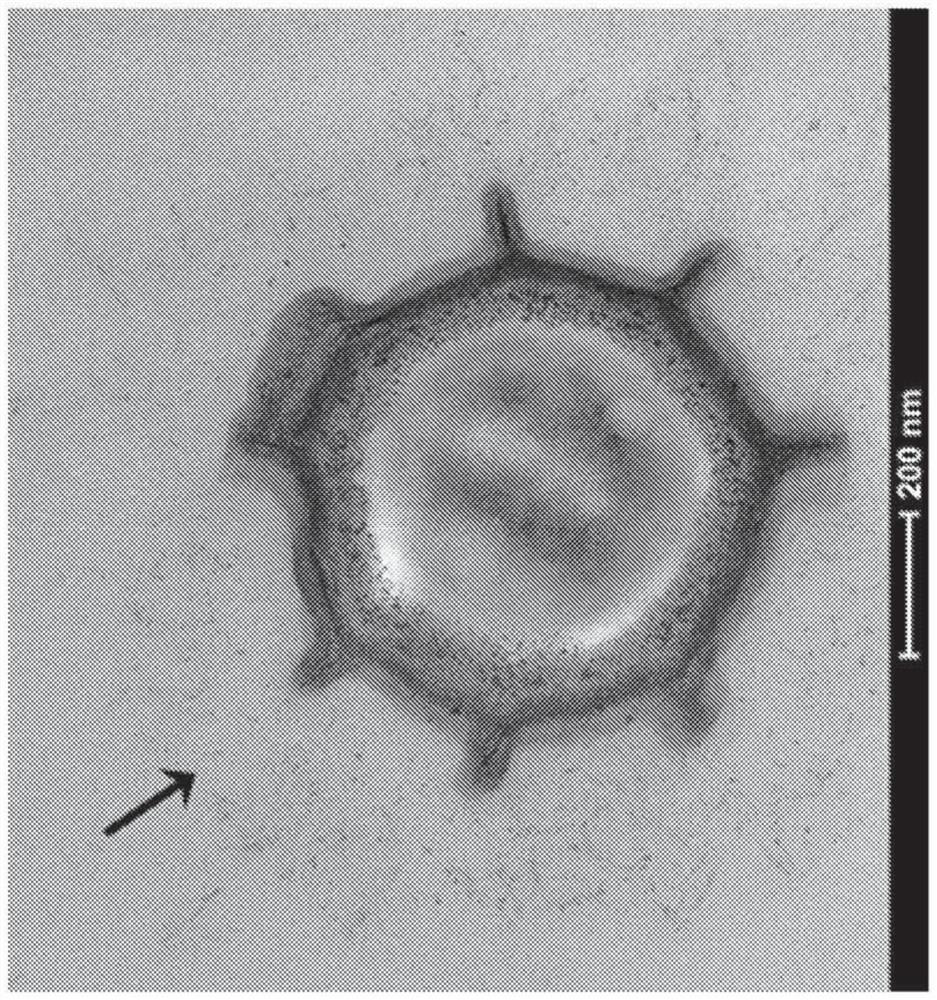
[0247] Search for ORFs containing collagen-like GXX repeats in the complete genome of Paenibacillus sp. NRRL B-50972. Collagen-like spore surface proteins were then observed by transmission electron microscopy ( figure 1 ). The presence of collagen-like spore surface proteins was also confirmed experimentally by mass spectrometry. Briefly, Paenibacillus NRRL B-50972 spores were trypsinized to remove surface proteins. Spores were removed by centrifugation, and the supernatant was analyzed by mass spectrometry to verify the presence of collagen-like spore surface proteins. This general protocol was used to identify endogenous Paenibacillus sp. NRRL B-50972 proteins with N-terminal targeting sequences identified by SEQ ID NO: 1-10. The same method can be used to identify spore surface proteins and corresponding N-terminal targeting sequences from Chlorobaci...
Embodiment 2
[0248] Example 2: General Protocol for the Preparation of Recombinant Paenibacillus Endospores Displaying Green Fluorescent Protein (GFP)
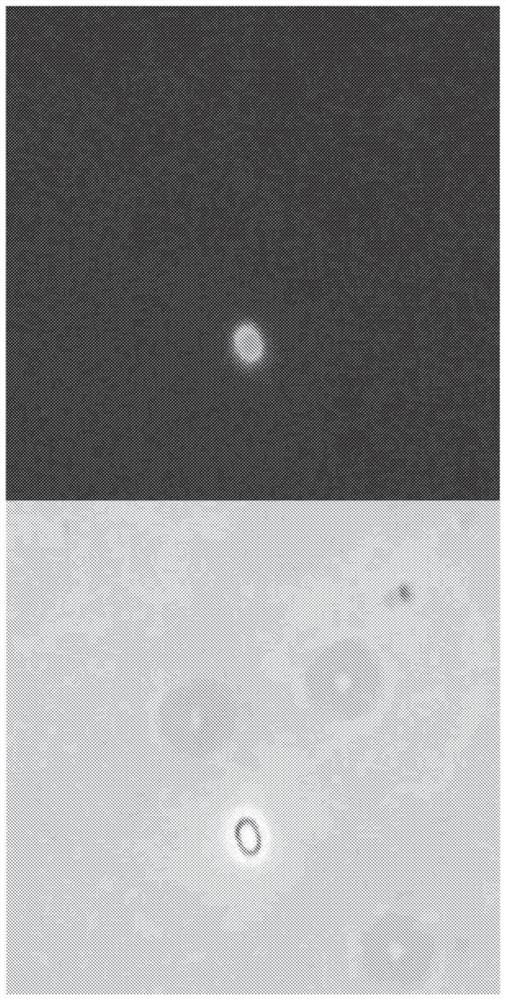
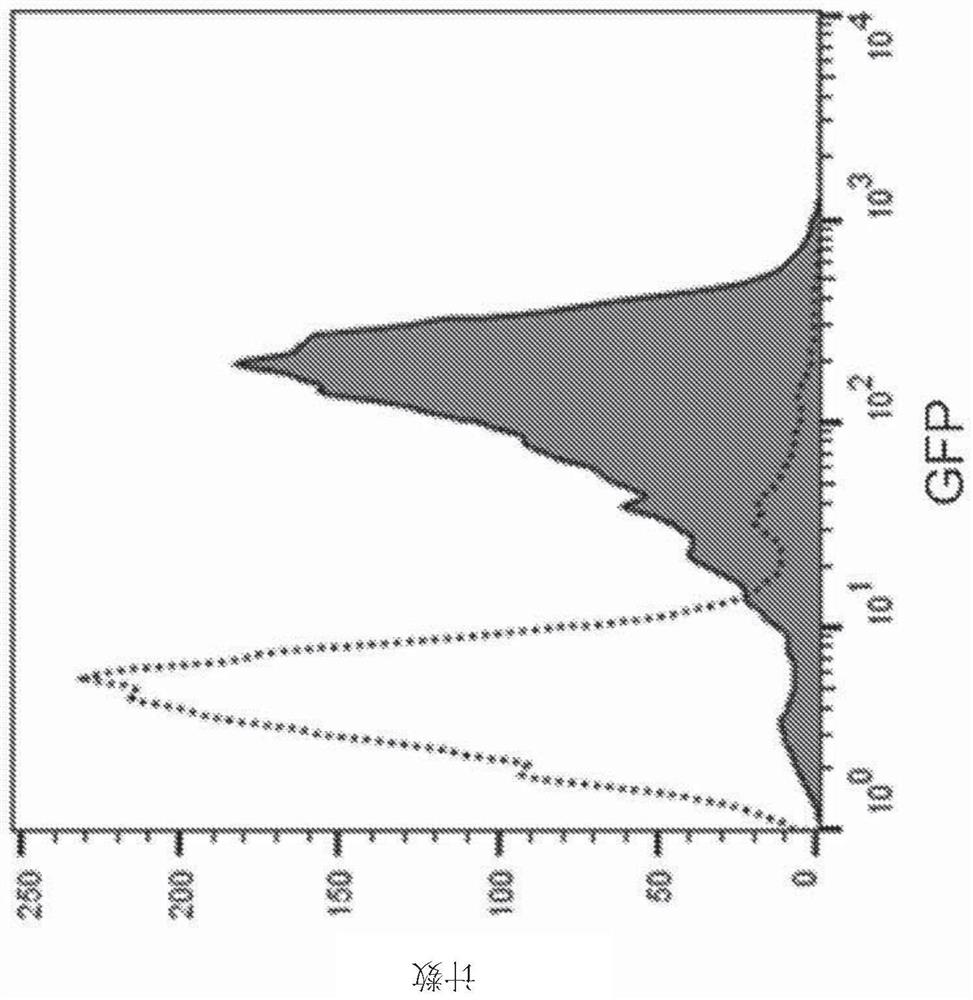
[0249] To generate the fusion construct, the gene encoding GFP was combined by gene synthesis with a DNA segment encoding the amino acids of the published N-terminal targeting sequence (SEQ ID NO: 1 ) of Paenibacillus sp. NRRL B-50972 (published in fused under the control of the native promoter of the N-terminal targeting sequence) and cloned into the E. coli / Paenibacillus shuttle vector pAP13. The resulting vector construct was introduced into Paenibacillus sp. NRRL B-50972. Correct transformants were then grown in Schaeffer's sporulation broth at 30°C until sporulation. Paenibacillus sp. NRRL B-50972 spores expressing the fusion construct were then examined by epifluorescence microscopy. GFP was visible on spores expressing the fusion construct ( Figure 2A ). Paenibacillus sp. NRRL B-50972 spores were also examined by flow cytometry...
Embodiment 3
[0250] Example 3: General protocol for making recombinant Paenibacillus endospores displaying any protein of interest
[0251] Paenibacillus cells (e.g., Paenibacillus sp. NRRL B-50972) can be cultured, transformed, and screened as described above in Example 2 to produce spores with N-terminal spore surface targeting sequences according to the present disclosure fusion constructs. Screening can be performed by mass spectrometry or any other biochemical or visual means known in the art (eg, the protein of interest can be tagged with GFP or other selection / screening tags). The N-terminal targeting sequence used to generate the fusion construct may comprise a polypeptide of any one of SEQ ID NO: 2, 4, 6, 8 or 10, 18, 20, 21, 22, 24, 26, 28, 30, or fragments or variants thereof. In some aspects, the N-terminal targeting sequence may comprise a residue having one or more residues corresponding to the pairwise alignment of SEQ ID NO: 2 and 8 ( image 3 ) sequence of the same resi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com