Transcript annotation method and method for screening long non-coding RNA and endogenous retrovirus source long non-coding RNA
A retrovirus, transcript technology, applied in the field of bioinformatics, can solve problems such as low throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1. RSCS Annotated Transcripts
[0041] This example uses mouse embryonic fibroblasts (MEFs) as an example to describe the method for annotating transcripts described in the present invention.
[0042] 1. Obtain the off-machine data of RNA sequencing and small RNA sequencing of MEFs and iPSCs from the first day to the eighth day of reprogramming (reprogramming) MEFs and iPSCs during the process of cell reprogramming.
[0043] 1. Use trim_galore (0.4.5) or cutadapt (1.18) software to de-join the off-machine data of RNA-seq and small RNA-seq to obtain clean data.
[0044] 2. Then use FastQC (v0.11.5) software to perform data quality control on the clean data obtained in step 1. The screening conditions are: 1) The sequencing quality score of each base is not less than 20; 2) The GC content of each sequence It conforms to a normal distribution, and the deviation does not exceed 15%; 3) the N content in the sequencing result does not exceed 5%; 4) the sequencing len...
Embodiment 2
[0053] Example 2. Method for screening long non-coding RNA
[0054] This example describes the screening of long non-coding RNAs using the annotation method in Example 1. The specific method is as follows:
[0055] 1. To annotate transcripts, refer to Step 1 in Example 1 for specific methods.
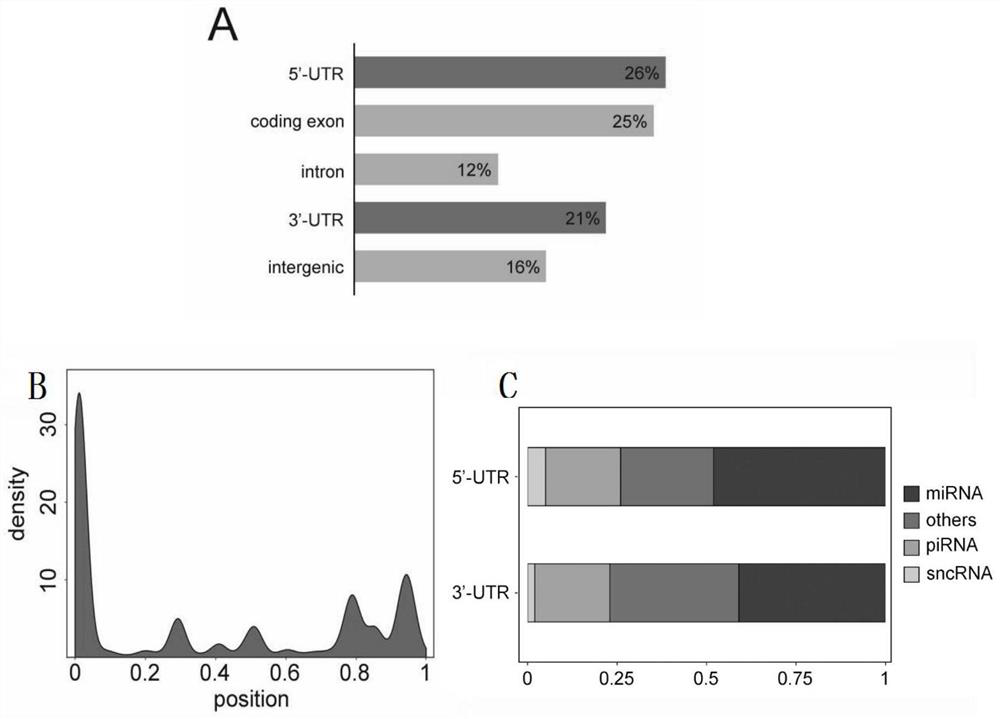
[0056] 2. Use CPC2 and CNCI software to predict the coding ability of the transcripts obtained in splicing step 1 at each time point of cell reprogramming. The results are as follows Figure 5 As shown in A, 13,072 long non-coding RNAs were obtained, accounting for 22.19% of the total transcripts; as Figure 5 As shown in B, there are 10,361 known long non-coding RNAs, accounting for 79.26%; 2,711 novel long non-coding RNAs, accounting for 20.74%. R language was used to compare and analyze the expression levels and coding abilities of long non-coding RNAs and coding genes in mefs and iPSCs. The results are as follows: Figure 5 As shown in C and D, the expression level and length of ...
Embodiment 3
[0057] Example 3. Method for screening endogenous retrovirus-derived long non-coding RNA
[0058] This embodiment describes a method for screening endogenous retrovirus-derived long non-coding RNA using the annotation method in Example 1, and the specific method is as follows:
[0059] 1. Screen out the encoded long non-coding RNA, refer to Example 2 for the specific method.
[0060] 2. Then use the bedtools interact software to select the long non-coding RNA within 5kb from the long non-coding RNA obtained in step 1 according to the position on the chromosome as the long non-coding RNA derived from the endogenous retrovirus. Non-coding RNAs (ERV-lncRNAs), the results are as follows Image 6 As shown, 40.8% of long non-coding RNAs contained TE (transposable element) sequences, of which 59.3% were long non-coding RNAs associated with endogenous retroviruses.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com