Primer, probe, kit and method for detecting HLA deletion type recurrence of patient
A deletion-type and kit-based technology, applied in the field of genetic engineering, can solve the problem of inability to benefit from lymphocyte infusion, and achieve the effects of easy interpretation of results, simple operation, and high type coverage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Example 1: Specificity of Primer Pairs and Probes for HLA Deletion Recurrence Detection
[0058] In this embodiment, primer pairs and probes are designed for specific HLA types, and the specificity of the primer pairs and probes is judged by the Ct value of the fluorescent quantitative PCR reaction.
[0059] In the present invention, samples with a Ct value of ≤30 are judged as positive, and samples with a Ct value of ≥40 or no amplification curve are judged as negative.
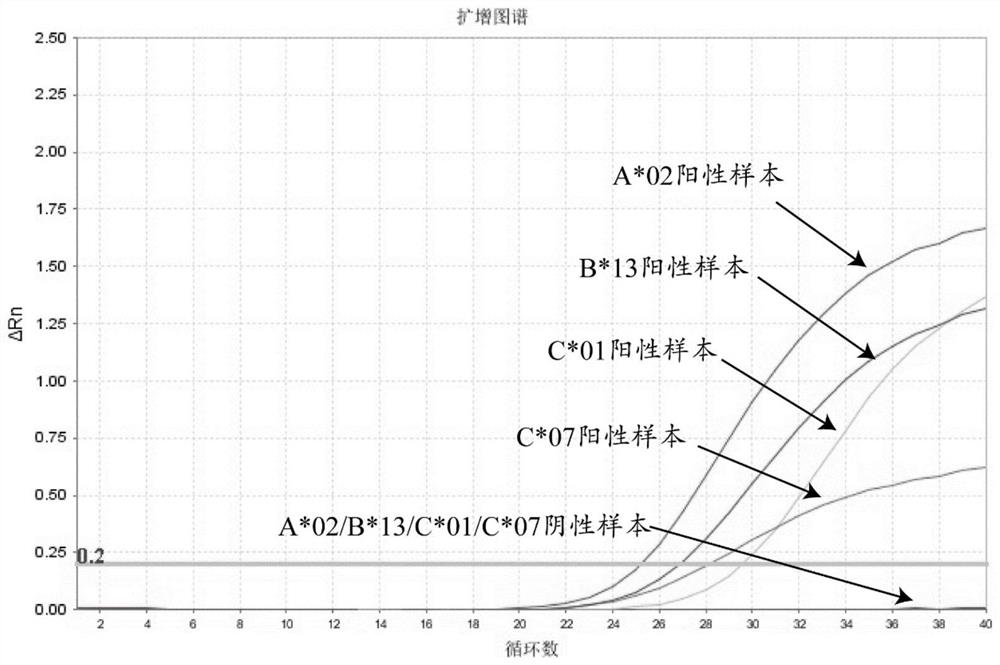
[0060] combined with figure 1 , from the amplification curves of positive and negative samples of specific HLA types A*02, B*13, C*01 and C*07, it can be seen that the Ct values of positive samples are 25.2, 26.9, 29.6 and 28.1 respectively, Negative specimens show no amplification. It shows that the above-mentioned types of primer pairs and probes have good specificity.
Embodiment 2
[0061] Example 2: Amplification Efficiency of HLA Deletion Recurrence Detection Primer Pairs and Probes
[0062] In this example, laboratory donor samples of HLA-A*02, B*13, C*01, and C*07 types detected by next-generation sequencing were selected as positive templates, and samples without this type were regarded as negative.
[0063] The positive and negative DNA samples were diluted to 15ng / μl, and the negative sample DNA was used to perform a 5-fold serial dilution of the positive sample. The positive samples (100%, 20%, 4%, 0.8%, 0.16%) were used for qPCR detection, and each sample was replicated three times, and the standard curve was drawn using the positive sample concentration and the average value of Ct.
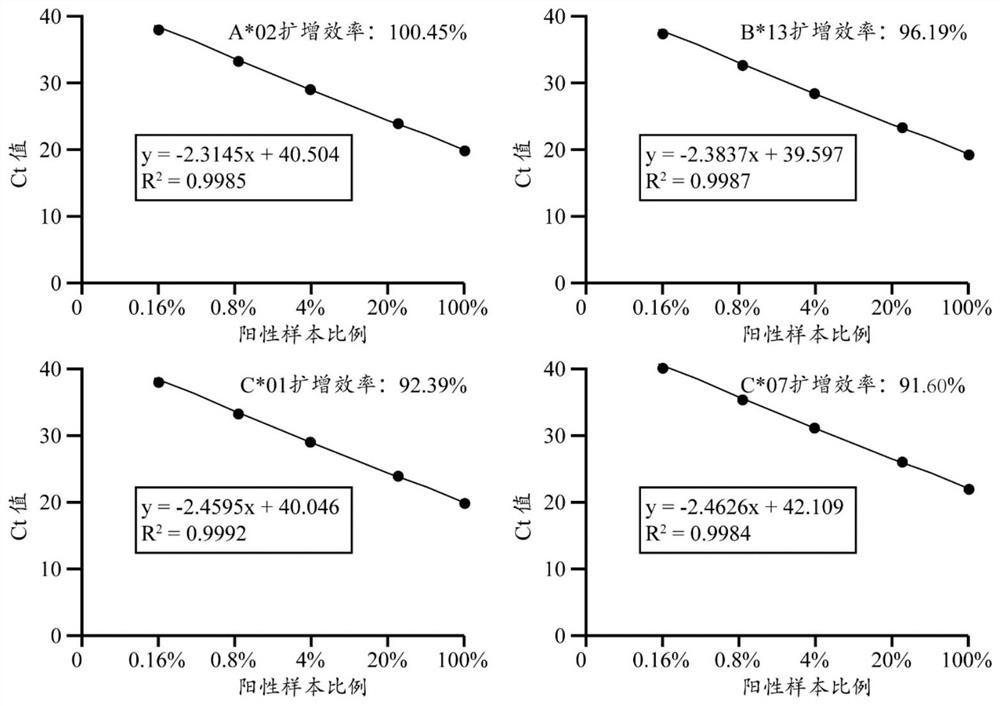
[0064] combined with figure 2 , it can be seen that the above site standard curve correlation coefficient R 2 >0.99, confirming that the primers amplified linearly within the range of 0.16%-100%.
[0065] by attaching figure 2 The slope of the amplification cu...
Embodiment 3
[0066] Example 3: Sensitivity of HLA Deletion Recurrence Detection Primer Pairs and Probes
[0067] In this example, HLA-A*01, A*02, A*11, A*23, A*25, B*13, B*35, B*38, C*01, C*02 detected by first-generation sequencing were selected. , C*03, C*04, C*05, C*07, DRB1*04, DRB1*12, DQB1*02, DQB1*04, DQB1*05, DPB1*01:01, DPB1*02:01, DPB1 *04:01 and DPB1*05:01 laboratory donor samples were taken as positive templates, and samples without this type were taken as negative.
[0068] The DNA concentrations of the above samples were all diluted to 15ng / μl, and the positive samples were diluted to 0.16% with the negative samples, and 2 batches were tested every day for 5 consecutive days.
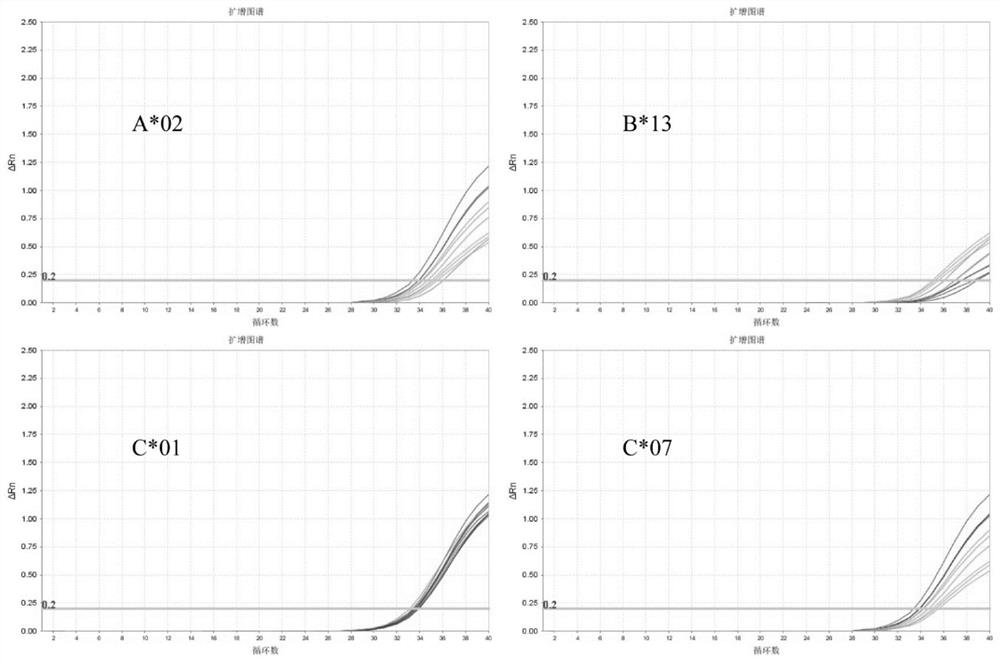
[0069] The detection results are shown in Table 5. The coefficients of variation of the above-mentioned type primer pairs and probes between batches and within batches of Ct values are all less than 5%, and the detection results of A*02, B*13, C*01 and C*07 are as follows: attached image 3 As sho...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com