A method for assembling multidomain protein structures based on interdomain residue contacts
An assembly method and domain protein technology, which can be used in bioinformatics, hybridization, instrumentation, etc., can solve problems such as low accuracy, and achieve the effect of improving prediction accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0042] The present invention will be further described below in conjunction with the accompanying drawings.
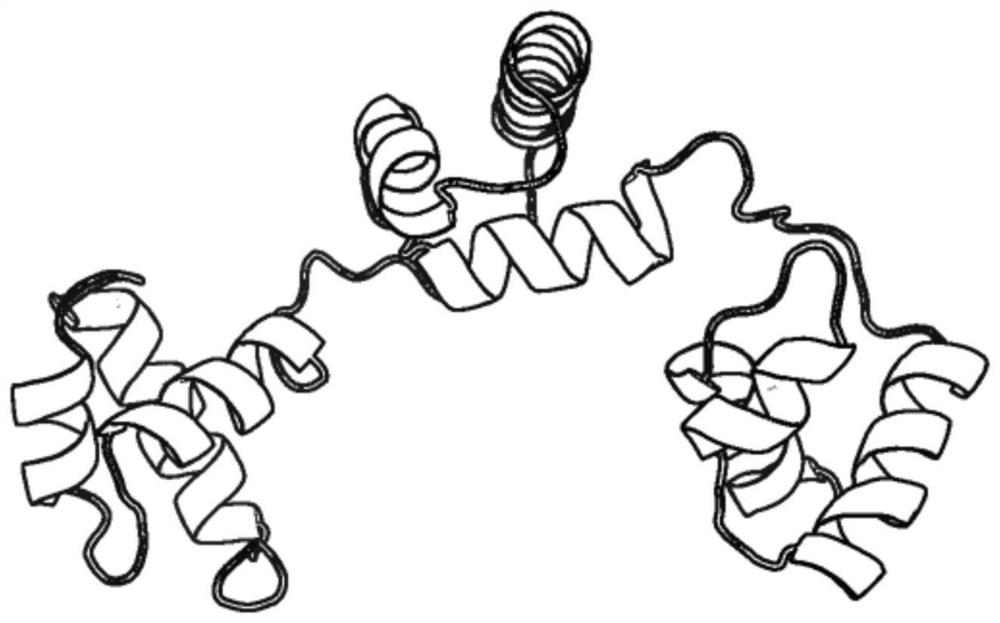
[0043] refer to figure 1 and figure 2, a multi-domain protein structure assembly method based on interdomain residue contacts, comprising the following steps:
[0044] 1) Given the three-dimensional structure and full-length sequence information of each domain of the protein to be assembled;
[0045] 2) Parameter setting: set the maximum number of iterations I max , conflict distance threshold d clash , the number of assembled templates T, the temperature length K, the contact energy depth d well ;
[0046] 3) Use the structural alignment tool TM-align (https: / / zhanglab.ccmb.med.umich.edu / TM-align / ) to each of the protein PDB library (http: / / www.rcsb.org / ) Multi-domain protein is scored, and the score of each template is the average value of TM-score (https: / / zhanglab.ccmb.med.umich.edu / TM-score / ) between each domain and the template, and the score is selected ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com