A dcl gene deletion mutant of Fusarium wilt of banana and its small RNA in race 4
A technology for banana withering and gene deletion, applied in DNA/RNA fragments, recombinant DNA technology, microorganisms, etc., can solve problems such as harm and threat, economic loss of banana industry and banana growers, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
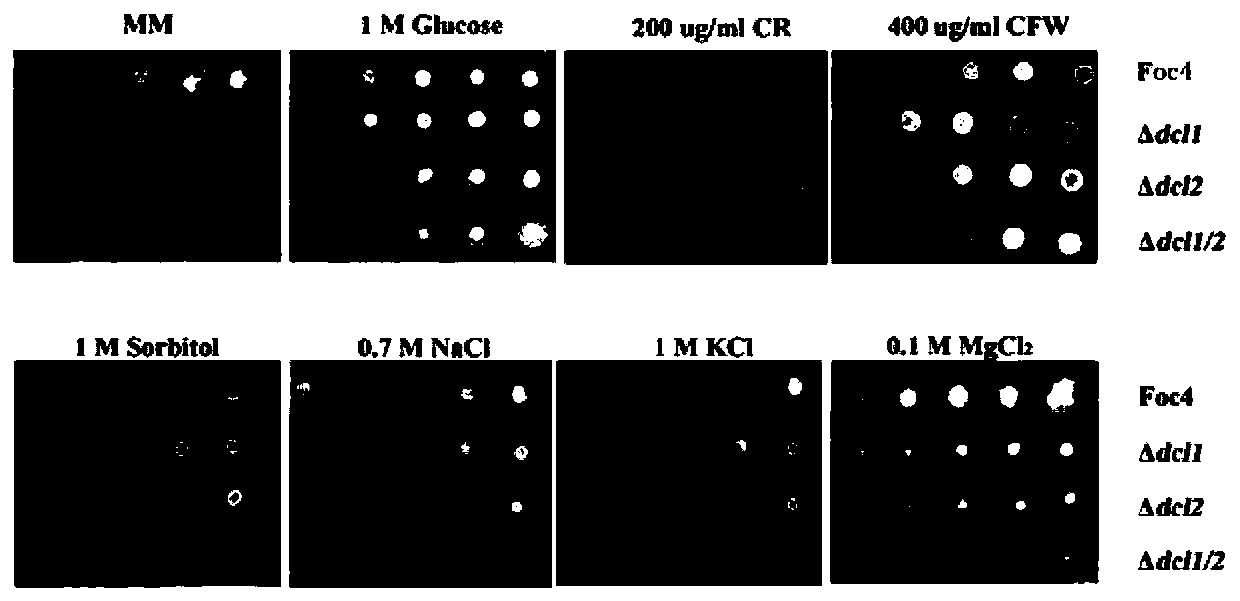
[0055] Example 1 Fusarium wilt Foc4 DCLs gene knockout
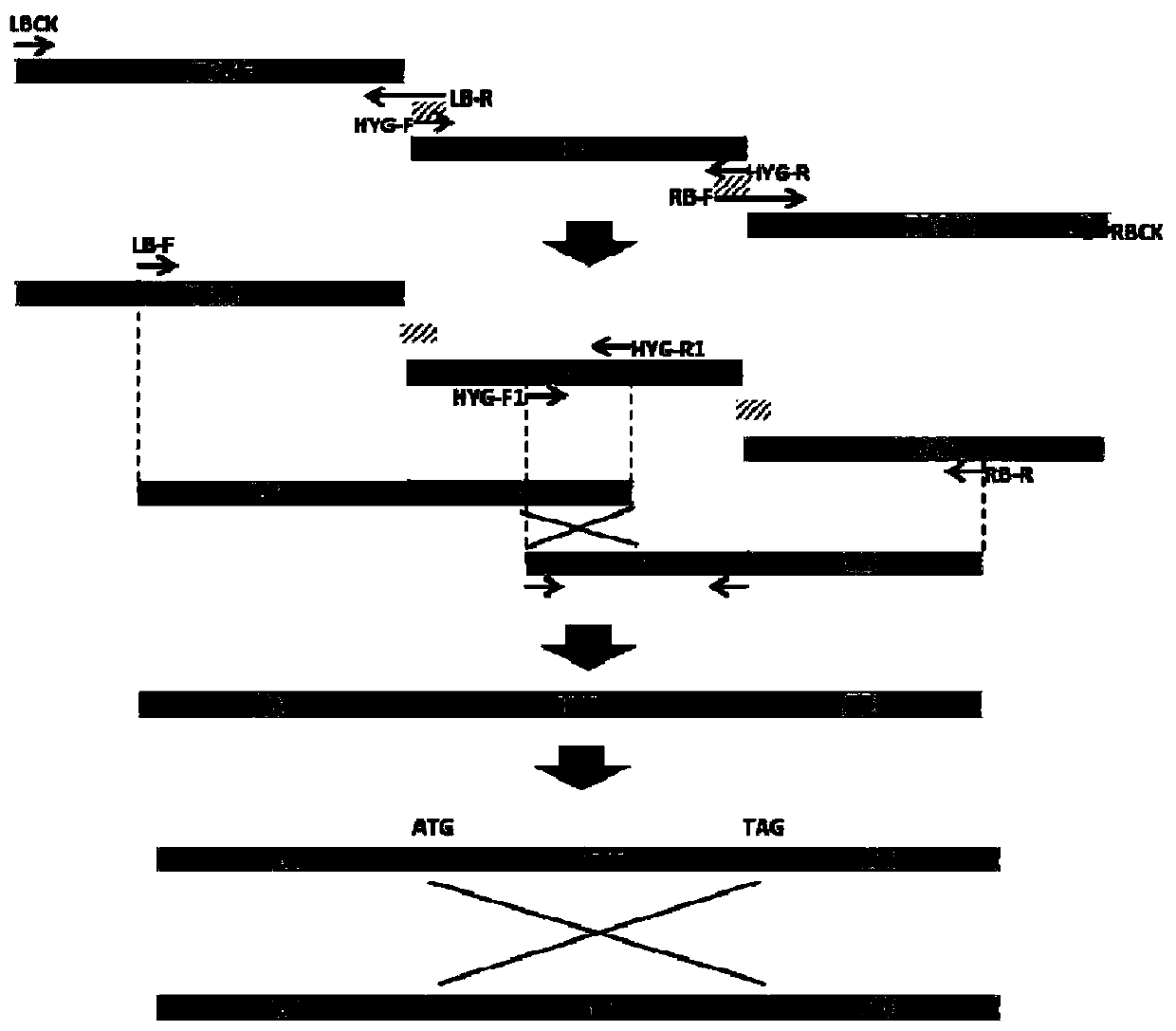
[0056] Fusarium wilt contains two DCL protein genes, DCL1 and DCL2, and the single gene knockout of Foc4 DCL adopts the principle of homologous replacement, replacing the DNA of the target gene Foc4 DCL1 or Foc4 DCL2 with the DNA fragment of the resistance gene hygromycin B (HPH) gene fragment, figure 1 It is a schematic diagram of the Split-PCR gene knockout method, and the primers used in the subsequent gene knockout steps are consistent with those in the schematic diagram.
Embodiment 2
[0057] Example 2 Construction of Foc4 DCL1 Gene Deletion Recombinant DNA Fragment
[0058] The split-PCR gene knockout method uses two rounds of PCR to amplify the recombinant DNA fragment, and the first round of PCR amplifies three fragments: the DNA sequence of the homology arm upstream of the target gene, the full-length sequence of the resistance gene, and the DNA of the homology arm downstream of the target gene sequence. In the second round of PCR, the DNA sequences of the upper and lower homology arms and the DNA of the resistance gene were mixed as PCR templates to amplify the upstream and downstream missing recombinant DNA fragments respectively. The gene sequence number of Foc4 DCL1 is Gene: FOIG_04851, referring to the genome sequence of NCBI GenBank: JH658277.1, primers are designed to amplify the homology arm gene sequence of the upper and lower reaches of the DCL1 gene ( figure 2 -A,B). details as follows:
[0059] The first round of PCR amplification:
[00...
Embodiment 3
[0069] Example 3 Construction of Foc4 DCL2 Gene Deletion Recombinant DNA Fragment
[0070] The gene knockout method of Foc4 DCL2 is the same as that of DCL1, and the primers and descriptions used are shown in Table 2. The gene sequence number of Foc4 DCL2 Gene: FOIG_04495, referring to GenBank: JH658276.1 genome sequence, designed primers to amplify the upper and lower homologous genome sequences of DCL2 gene ( figure 2 -C,D). The detailed operation method is as follows:
[0071] The first round of PCR amplification:
[0072] Left LB: DCL2-LBCK and DCL2-HPH-LB-R are primers, FOC4 genomic DNA is a template, and the size of the amplified product is 1957bp;
[0073] RB at the right end: DCL2-HPH-RB-F and DCL2-RBCK are primers, FOC4 genomic DNA is a template, and the size of the amplified product is 1837bp;
[0074] HPH fragment: HYG-F and HYG-R are primers, vector plasmid DNA is a template, and the size of the amplified product is 1376bp;
[0075] The second round of PCR am...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com