Multi-omics cancer data integrating and analyzing method based on similarity fusion
A similarity fusion and data integration technology, applied in the field of bioinformatics, can solve problems such as high dimensionality, high integration methods, and large noise
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0059] The present invention will be further described below in conjunction with specific examples.
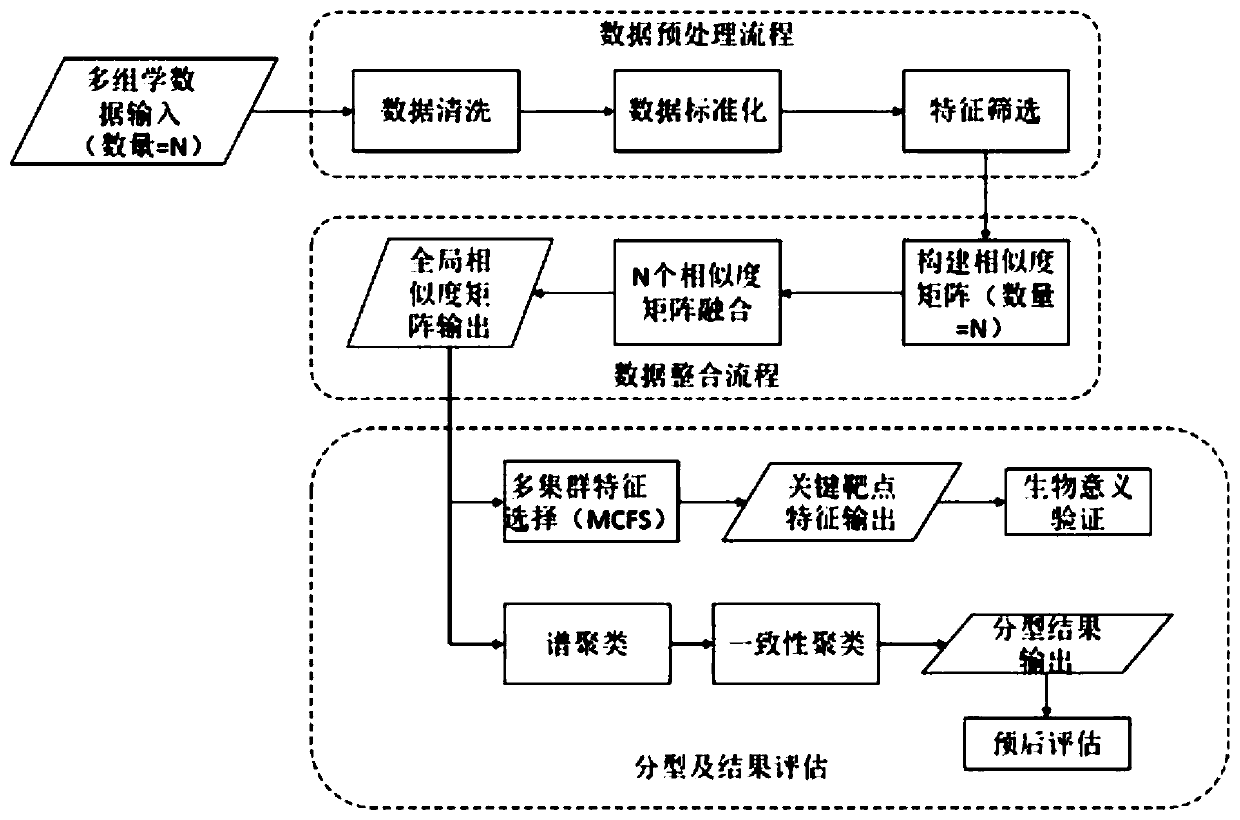
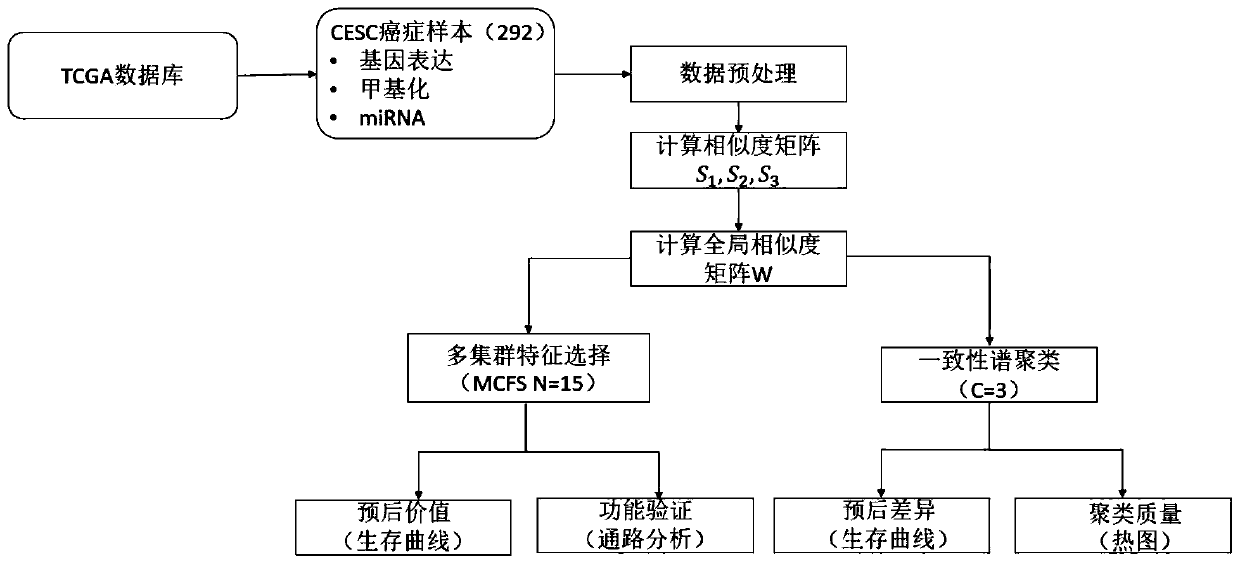
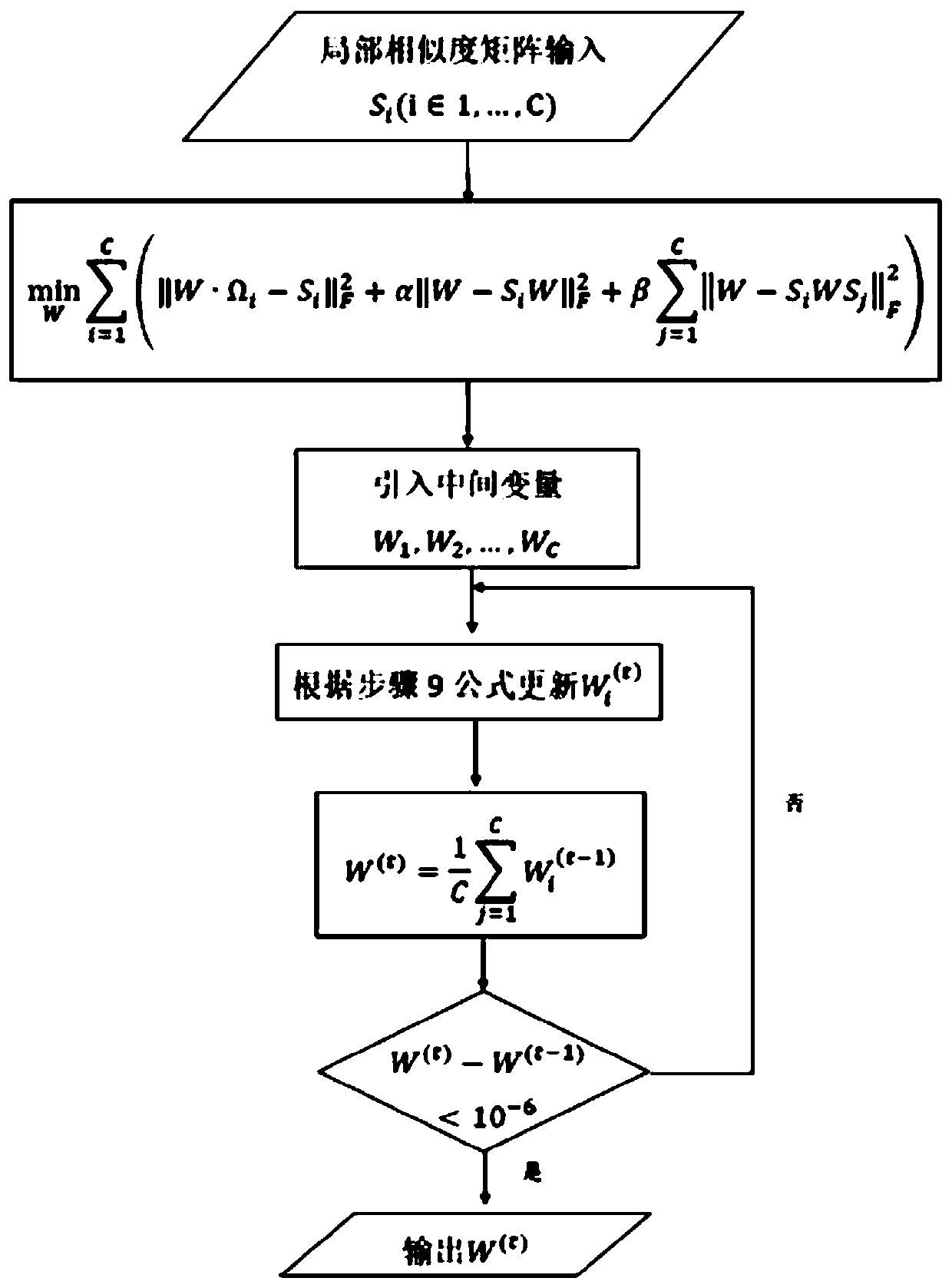
[0060] This embodiment uses the cervical cancer project (CESC) data in the public cancer data set TCGA to evaluate the method of the present invention, and uses different types of patients as evaluation indicators. The outline design of this example is as follows figure 1 As shown, the implementation process is as follows figure 2 As shown, the specific process of realizing the multi-omics cancer data integration analysis method based on similarity fusion based on similarity fusion is as follows:
[0061] Step 1. Obtain the gene expression data, methylation data, and miRNA data of the same batch of samples of the cervical cancer project in the TCGA database. In this example, the data of 292 cervical cancer patients were initially collected.
[0062] Step 2, data cleaning: process the null values in the data, delete more than 20% of the samples or features directly, and use...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com