Mds to aml transition and prediction methods therefor
A prediction model and transcriptome technology, applied in the field of transition from MDS to AML and its prediction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
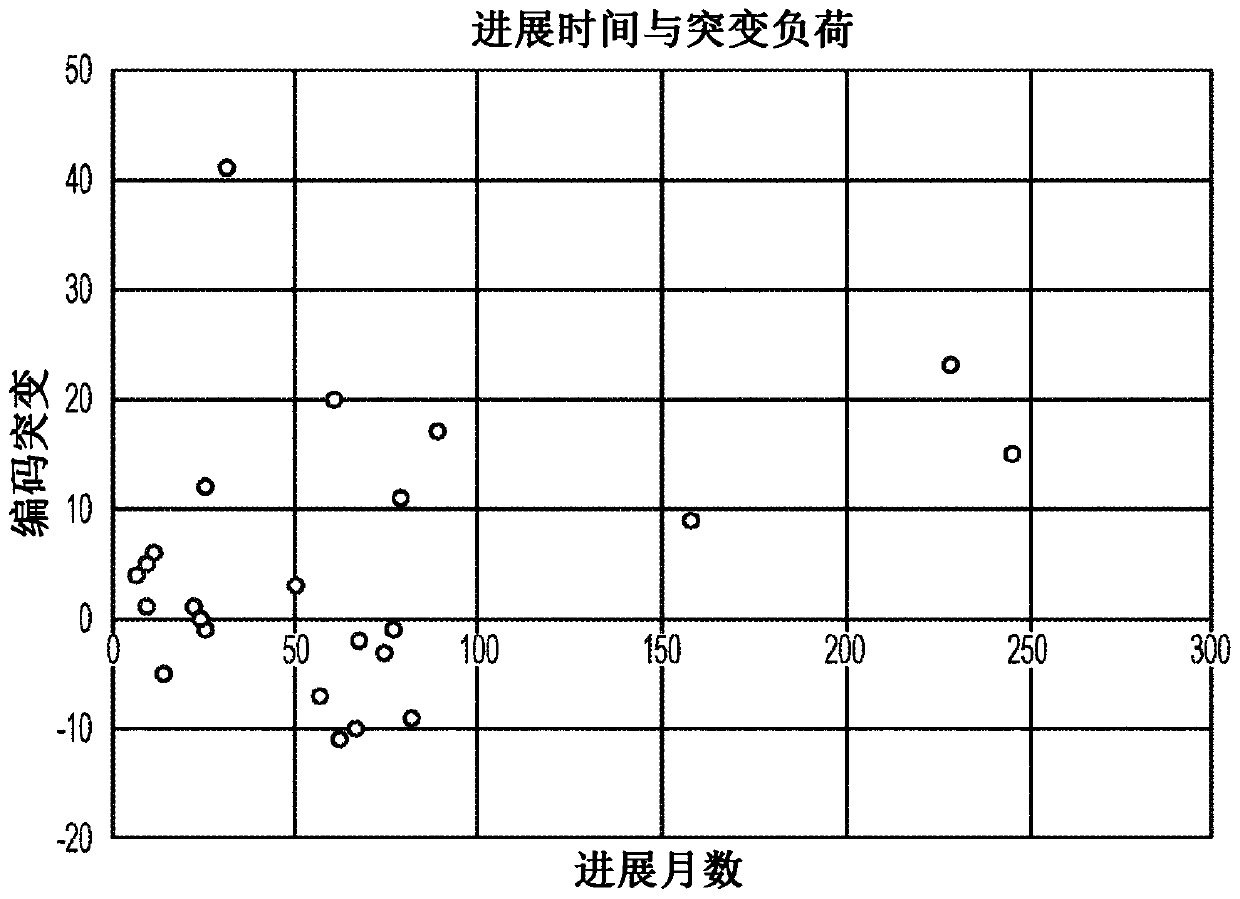
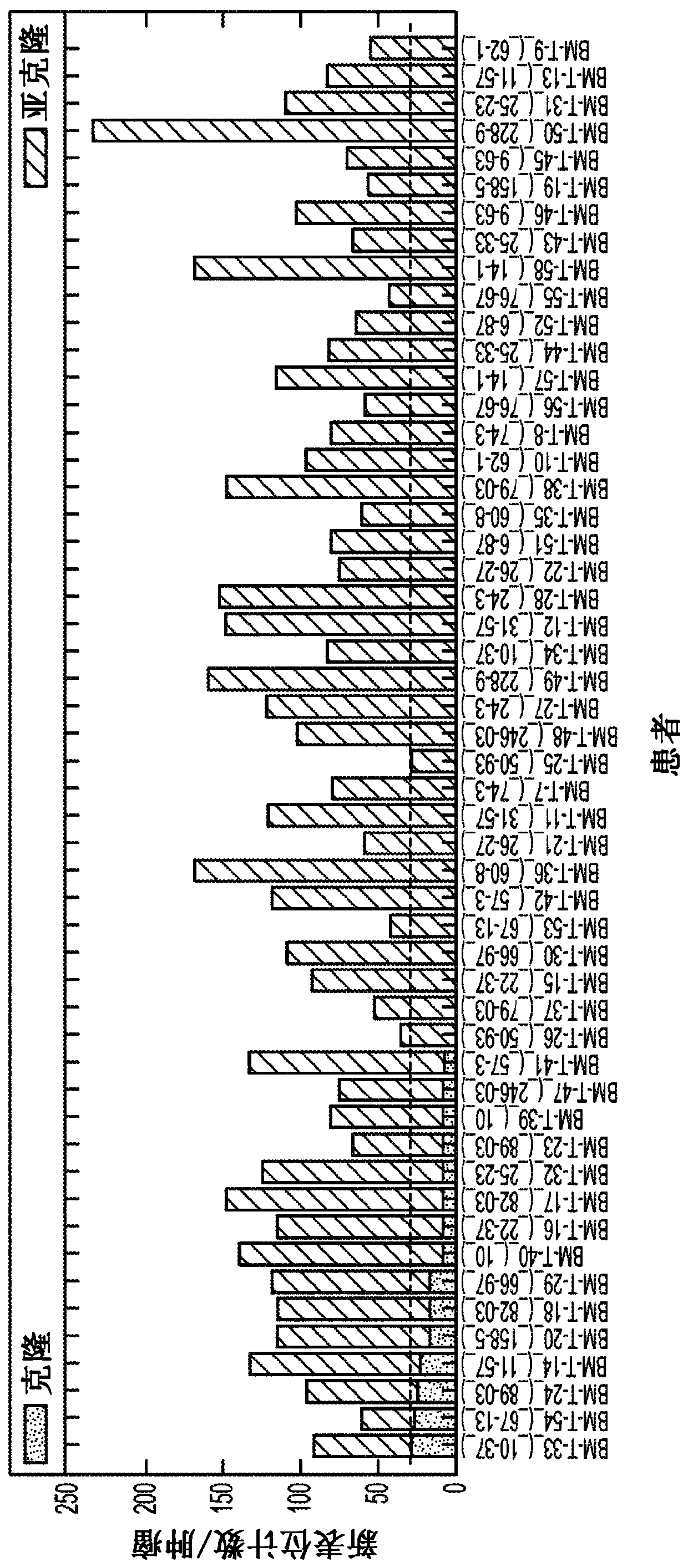
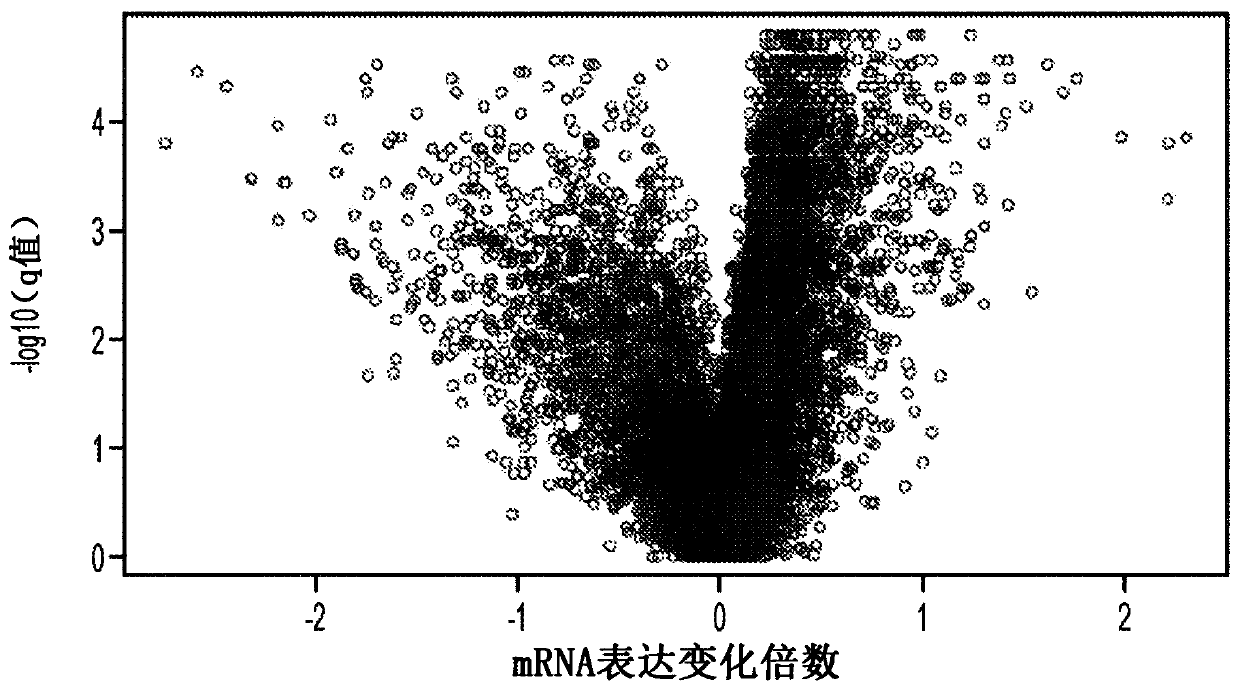
[0031] In a first attempt to identify predictive markers for the progression of MDS to AML, the inventors compared patient data with different progression times and mutational burdens, particularly of protein-coding genetic sequences. Omics analysis was performed using whole-genome sequencing of MDS and AML cells from the same patient and incremental localization-guided simultaneous alignment using BAMBAM, such as described in US9721062. figure 1 Exemplary results of this analysis are depicted. Notably, the median mutational change was approximately +2.5 coding mutations in the patient population with progression time <38 months, whereas the median mutational change in the patient population with progression time greater than 38 months and less than 80 months is about -2.0 coding mutations. On the other hand, among patients with a time to progression of more than 80 months, the median mutational change was approximately +15.0 coding mutations. While this increase at least a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com