Breast cancer occurrence related characteristic gene screening method
A screening method and related feature technology, applied in the field of bioinformatics, can solve the problems of missed selection of important genes, inability to fully retain characteristic genes, lack of effectiveness of complex collinearity processing, etc., and achieve the effect of high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
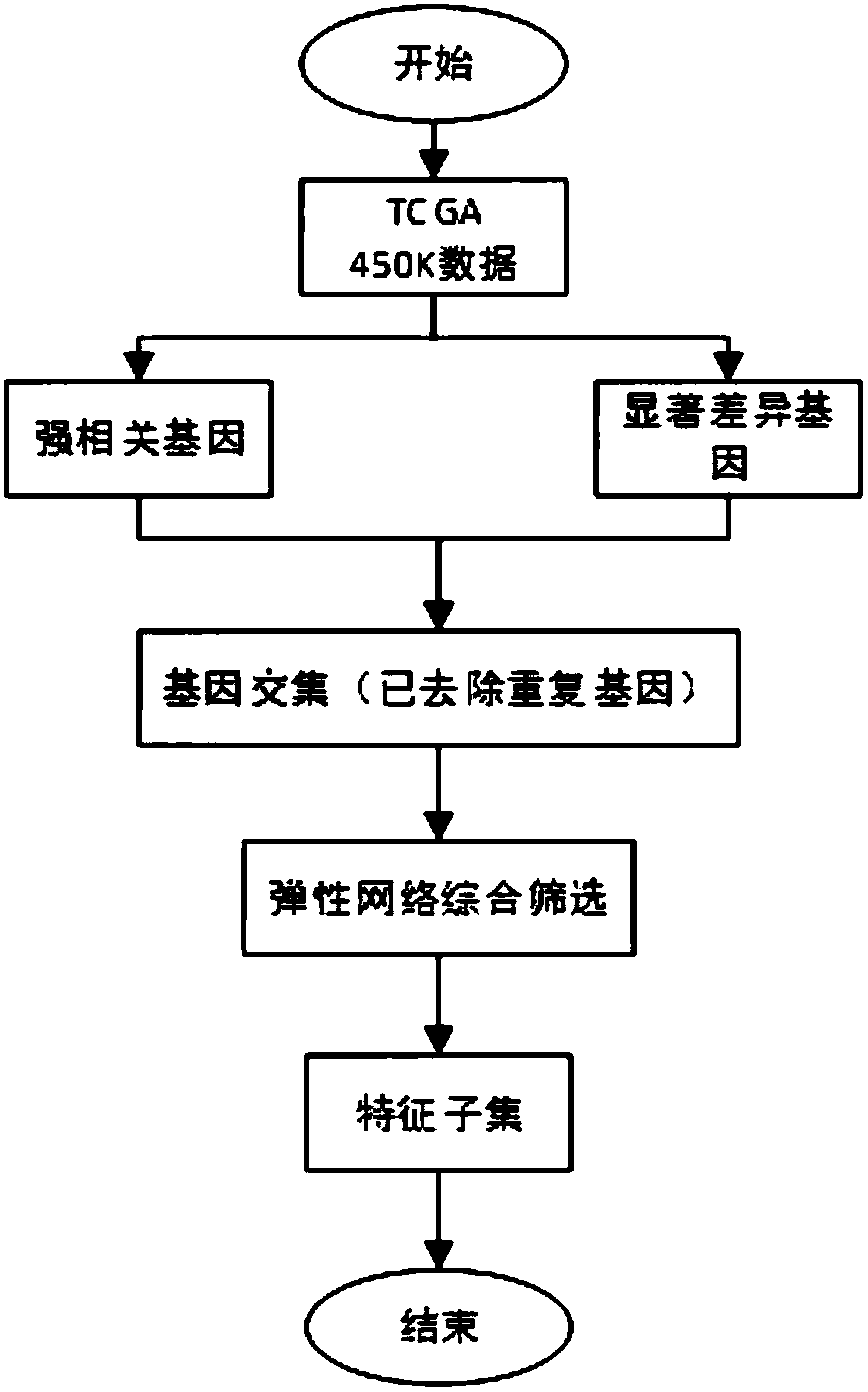
[0014] Such as figure 1 As shown, the present invention provides a method for screening characteristic genes related to breast cancer occurrence. Based on the breast cancer data in the TCGA cancer genomics database, multiple screening methods are used to screen the characteristic genes for cancer occurrence and used for classification by classification models.
[0015] Preferably, the multiple screening method comprehensively performs multi-step screening on the whole genome by combining correlation screening, significant difference screening and elastic network screening.
[0016] Aspects of the method are described in detail below in conjunction with the data.
[0017] 1. Material selection and data processing
[0018] The present invention selects Illumina Infinium Human Methylation 450K, Illumina Infinium Human Methylation 27K two test platforms breast cancer methylation data and IlluminaHi Seq 2000RNA Sequencing Version 2 test platform gene expression data in the TCGA pu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com