Methods of isolating t cell receptors having antigenic specificity for a cancer-specific mutation
A specific and antigenic technology, applied in the direction of cancer antigen components, genetically modified cells, receptors/cell surface antigens/cell surface determinants, etc., can solve the problems of difficult identification of TCR and patient isolation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0124] This example demonstrates a method for identifying one or more genes in the nucleic acid of a patient's cancer cells, each gene containing a cancer-specific mutation encoding a mutated amino acid sequence.
[0125]A 43-year-old woman (Patient (Pt.) 3737) with extensively metastatic cholangiocarcinoma who had undergone multiple chemotherapy regimens was enrolled in TIL-based adoptive cell therapy (ACT ) scheme. The clinical characteristics of patient 3737 are shown in Table 1.
[0126] Table 1
[0127]
[0128] *Acquisition sites for TIL generation and whole exome sequencing
[0129] + Functional status: ECOG, Eastern Cooperative Oncology Group
[0130] Lung metastases were excised and used as a source for whole-exome sequencing and generation of therapeutic T cells. Table 2 shows the somatic mutations identified by whole exome sequencing of metastatic pulmonary nodules from patient 3737. Tumor nodules were estimated to be approximately 70% of the tumor by pathol...
Embodiment 2
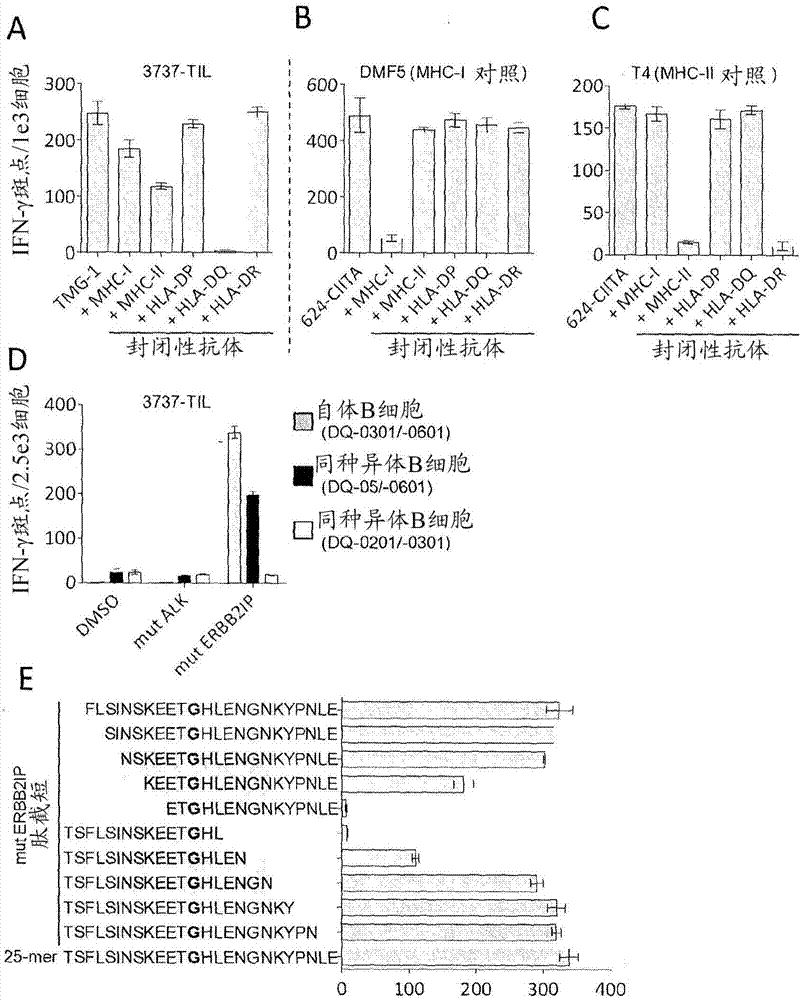
[0135] This example demonstrates the method of: inducing the patient's autologous APCs to present the mutated amino acid sequence; co-cultivating the patient's autologous T cell population with the autologous APCs presenting the mutated amino acid sequence; and selecting the autologous T cells that: (a ) is co-cultured with autologous APCs presenting the mutated amino acid sequence, and (b) is antigen specific for the mutated amino acid sequence presented in the context of MHC molecules expressed by the patient.
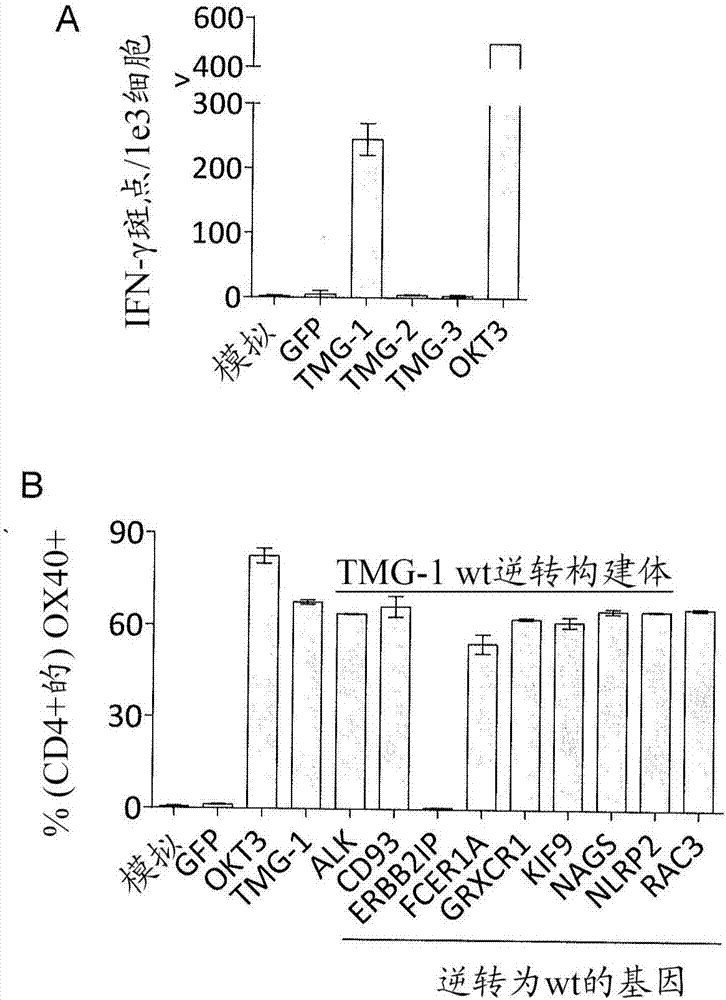
[0136] For each mutation identified in Example 1, a minigene construct was designed encoding the mutated amino acids flanked on each side by 12 amino acids from the endogenous protein. Multiple minigenes were synthesized in tandem to generate tandem minigene (TMG) constructs (Table 3). In Table 3, underlines indicate mutated amino acids and novel sequences encoded by point mutations or nucleotide insertions or deletions. For the splice site donor mutations (HLA-DOA ...
Embodiment 3
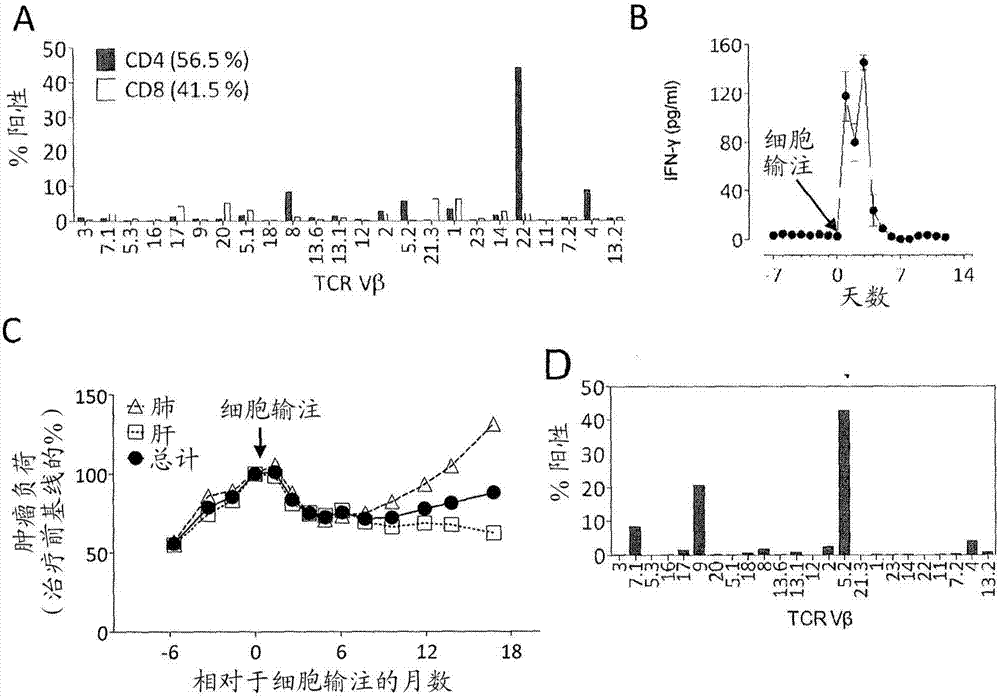
[0150] This example demonstrates that autologous open repertoire peripheral blood T cells genetically engineered with the TCR-Vβ22 chain (matched to its α chain) of ERBB2IP-specific CD4+ T cells identified in Example 2, are endowed with the ability to target the mutant Specific reactivity of ERBB2IP peptides.
[0151] The clonality of the mutant ERBB2IP-specific CD4+ T cells identified in Example 2 was characterized by sorting them after antigen-specific activation using OX40 as an activation marker. These cells were then bulk expanded and cloned by limiting dilution. Flow cytometry-based measurements of overall TCR-Vβ indicated that the overall expanded population was greater than 95% Vβ22+, and 10 / 11 clones assessed were pure Vβ22+. TCR sequence analysis revealed identical TCRβV-D-J sequences in 6 / 6 Vβ22+ clones tested (Table 5), suggesting that the majority of ERBB2IP-mutation reactive T cells contained a predominant Vβ22+ T cell clone.
[0152] table 5
[0153]
[01...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com