Adapters for Next Generation Sequencing
A next-generation sequencing library and sequence technology, which is applied in the determination/testing of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., can solve problems such as unrecognized and hindered research fields
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
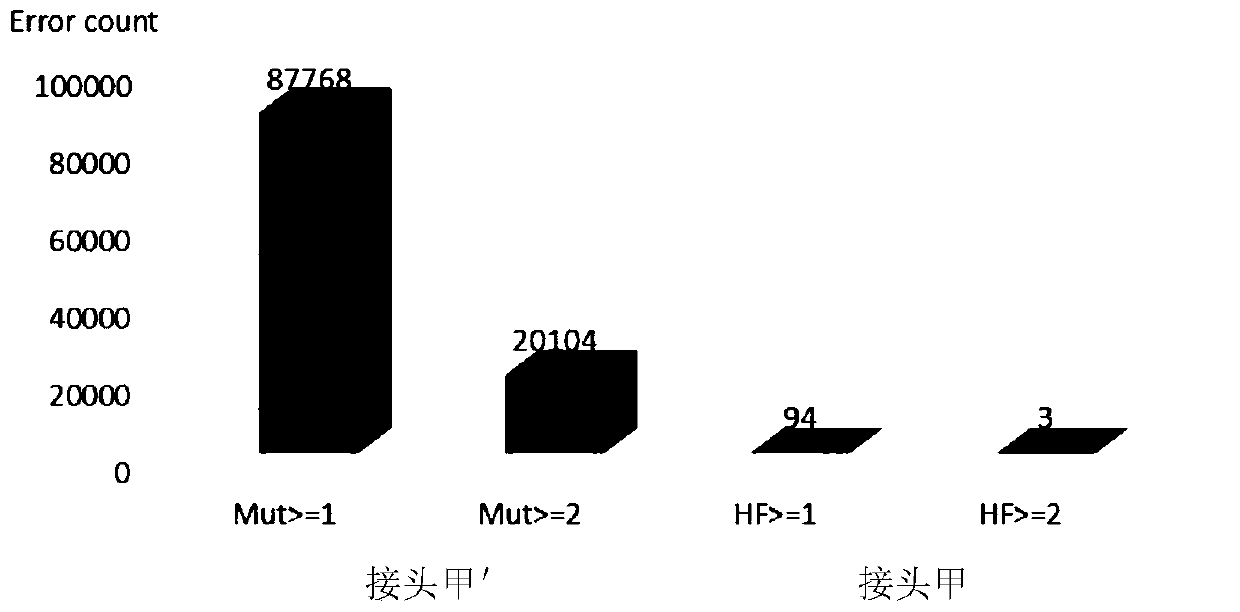
[0083] Example 1. A complete set of reagents used as adapters can improve the accuracy of high-throughput sequencing
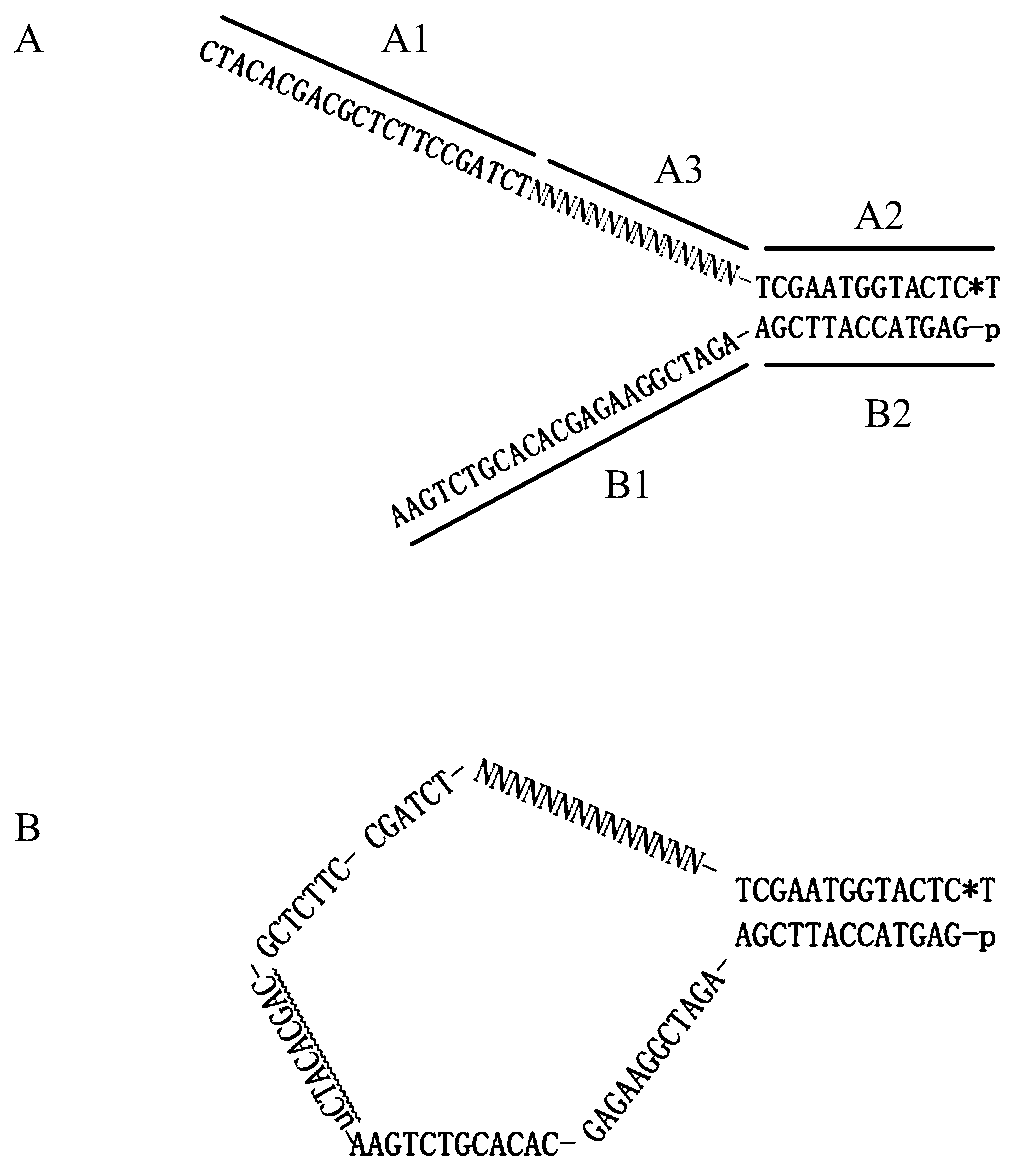
[0084] This embodiment provides a kit of reagents for next-generation sequencing as an adapter, which consists of two single-stranded DNAs named A strand and B strand;
[0085] The A chain is shown in formula (I) from the 5' end to the 3' end:
[0086] A1-A3-A2(I);
[0087] Each nucleotide of A3 is A, T, C or G, that is, A3 is a random sequence;
[0088] The B chain is shown in formula (II) from the 3' end to the 5' end:
[0089] B1-B2(II);
[0090] A2 and B2 are complementary; A1 and B1 are not complementary;
[0091] A3 is not complementary to both B1 and A1;
[0092] The sequences of A1 and A2 are different; the sequences of B1 and B2 are different;
[0093] The 3' terminal nucleotide in A2 is a thionucleotide, that is, the oxygen atom with a double bond on the phosphoric acid in the 3' terminal nucleotide is replaced by a sulfur atom; the 5' terminal...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com