Method for constructing QTL positioned new-group-linkage F2 group
A group and linkage technology, which is applied in modern biological science research and many research fields, can solve problems such as inestimability, difficulty in continuous research, and limited number of QTL mapping, so as to increase accuracy and reduce the loss of the same gene in RILS risk effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
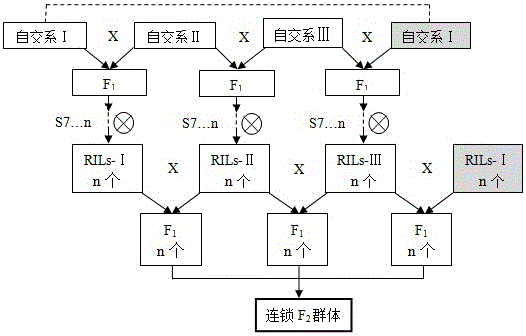
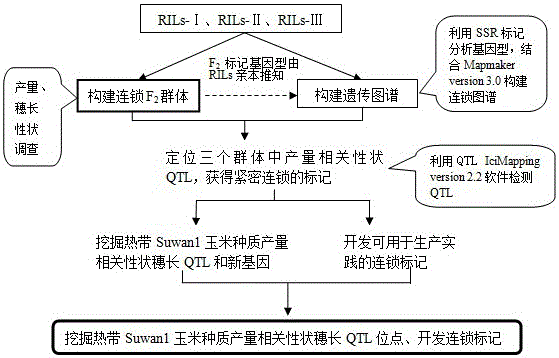
[0046] chain F 2 The group carried out research on gene mining and QTL mapping of excellent ear length related traits of tropical Suwan1 maize germplasm.
[0047] 1. Research materials and group construction
[0048] (1) Three maize inbred lines: Yunnan maize breeding backbone inbred line YML46 is derived from tropical Suwan1 maize germplasm, CML172 is derived from CIMMYT subtropical maize inbred line, and Ye 107 is derived from my country's temperate zone backbone maize inbred line.
[0049] Maize inbred line YML46 was published in Yao et al., Diallelanalysis models: a comparison of certain genetic statistics, CropSci., 2013, 53:1481-1490, CML172 was published in Yao Wenhua, etc., using SSR markers and yield to divide 27 maize inbred lines into heterosis groups, Maize Science , 2009, 17(1): 54-58 published, Ye 107 in Teng Wentao et al., Analysis of Chinese maize heterosis groups and their pattern changes in the past ten years, Chinese Agricultural Sciences, 2004, 37(12): 180...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com