Agaricus bisporus SSR molecular marker specific primer system and application thereof
A molecular marker and specific primer technology, applied in the field of molecular biology, can solve the problem of the same name of Agaricus bisporus, the same name of foreign body and so on
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1 Development of Agaricus bisporus EST-SSR marker primers
[0028] 1. Design and synthesis of SSR primers:
[0029] Search and download from the JGI database of Agaricus bisporus H97 genome data (version2.0) (http: / / genome.jgi-psf.org / .), using MISA (http: / / pgrc.ipk-gatersleben.de / tools .php) to search for SSR, the search criteria are: the minimum number of repetitions of two, three, four, five, and hexanucleotides is greater than or equal to 5 times.
[0030] Primers were designed in sequences containing SSR sites using the primer design software Primer3.0. SSR primer setting parameters are: (1) GC content is 45%-60%; (2) annealing temperature is 60°C; (3) expected fragment length is between 150-300bp; (4) primer length is 18-27bp between.
[0031] 2. Screening of SSR primers
[0032] (1) Extract the total DNA of the sample to be identified using Kangwei Century's genome extraction kit;
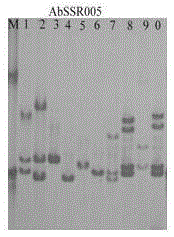
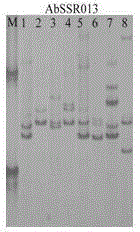
[0033] (2) Use 10 random Agaricus bisporus strains to conduct prelim...
Embodiment 2
[0045] Example 2 Application of Agaricus bisporus SSR marker primers in species identification
[0046] Select 10 pairs of SSR molecular markers with polymorphism and specificity to construct electronic molecular ID cards for the different strains screened above. Specific steps are as follows:
[0047] 1) The PCR products of 59 strains of each primer were also subjected to 8% polyacrylamide gel electrophoresis in step (3), and then the different electrophoresis band patterns were counted to determine whether the 59 strains were the same strain. The results showed that there were 22 specific strains among the 59 strains, and the remaining 37 strains were divided into 7 categories:
[0048] The first category: the same strain as CCMJA29 is CCMJA9, CCMJA21;
[0049] The second category: the same strain as CCMJA33 is CCMJA6;
[0050] The third category: the same strains as CCMJA22 are CCMJA1, CCMJA2, CCMJA11, CCMJA13, CCMJA14, CCMJA36;
[0051] The fourth category: the same st...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com