A molecular marker method for predicting and identifying superovulation traits in sheep and its molecular marker primers
A superovulation and molecular marker technology, applied in the directions of biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc. Reduced ovulation efficiency and other problems, to achieve the effect of increasing the number of eggs retrieved, reducing variation, and reducing the cost of testing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
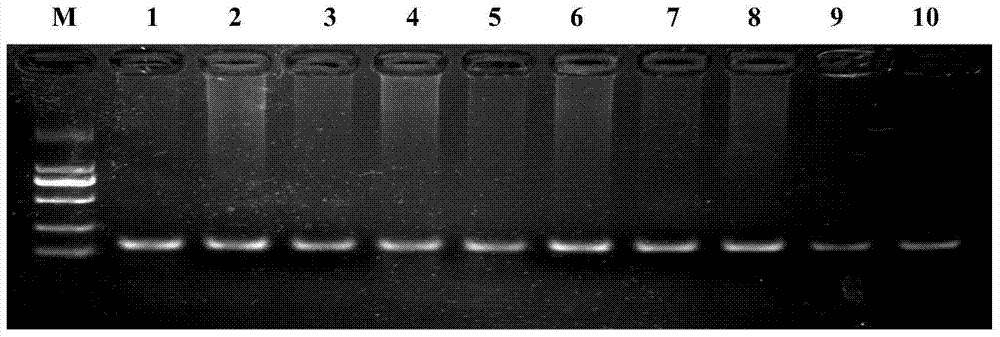
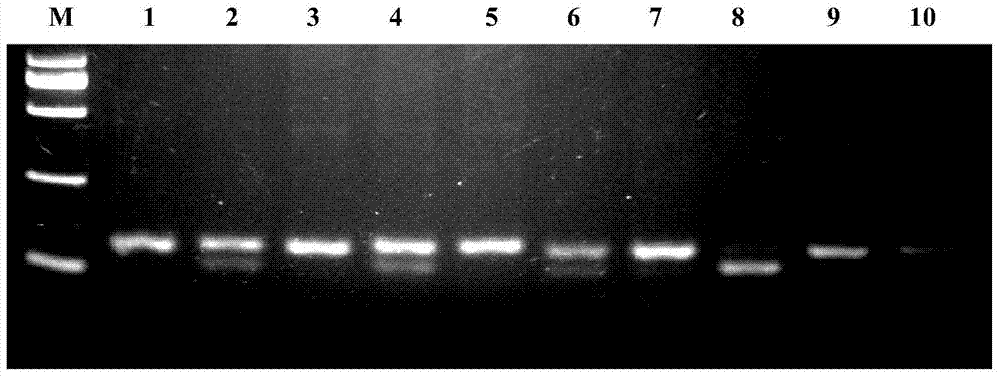
Image
Examples
specific Embodiment approach 1
[0021] Specific embodiment 1: A molecular marker primer for predicting and identifying superovulation traits of sheep according to this embodiment, the molecular marker primers are FSHRPF and FSHRPR, the sequence of FSHRPF is shown in the sequence table Seq ID No: 1, and the sequence of FSHRPR As shown in the sequence table Seq ID No: 2.
specific Embodiment approach 2
[0022] Specific embodiment 2: The difference between this embodiment and specific embodiment 1 is that the FSHRPF primers in the molecular marker primers are designed in the following way: the CATC is designed on the FSHRPF primer sequence by using the single base mismatching method of the primers. Bases, shielding the Sau3AI interference restriction site on the genome in the primer region, and only retaining the restriction site for C-365T mutation detection. Others are the same as in the first embodiment.
specific Embodiment approach 3
[0023] Specific embodiment 3: In this embodiment, the molecular marker method uses the C>T mutation site of c.-365 in the 5' regulatory region of the sheep FSHR gene for typing, so as to predict and identify the superovulation traits of sheep.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com