Method for cell lysis in rt‑PCR reaction buffer
A reaction buffer, RT-PCR technology, applied in the field of RNA expression analysis, can solve problems such as material loss
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
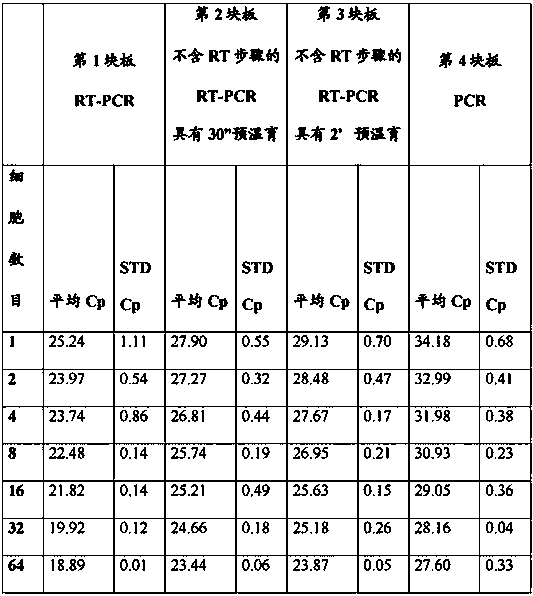
[0097] qPCR for amplification of GAPDH gene and RPLI13A gene from sorted mouse hybridoma cells
[0098] A defined number of mouse hybridoma cells were deposited into separate wells of a 96-well microtiter plate using a cell sorter (Beckton Dickinson, FACS Aria I) in such a way that the liquid beam was always directed into the center of the well. However, due to the underlying technology of the cell sorter, it cannot be ruled out that a small percentage of sorted particles are not intact whole cells but cell fragments. Hence, hereinafter, the amount of sorted material will be referred to as cell equivalent.
[0099] Cells sorted as published were dispensed according to the pipetting protocol described below into 96-well microtiter plates (Roche AppliedScience cat. no.: 04 729 692 001 ) designed for use with the LC480 Real-time PCR instrument (Roche Applied Science cat. no.: 05 015 278 001 ) Inside:
[0100] Columns 1-4 1 cell equivalent / well
[0101] 2 cell equivalents / well ...
Embodiment 2
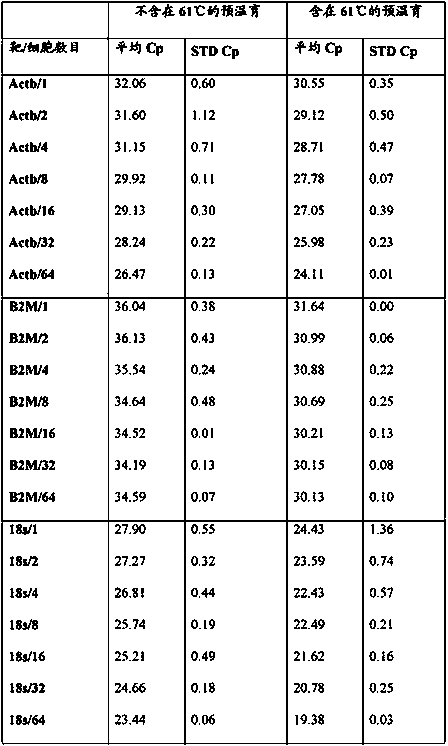
[0131] qPCR for amplification of the Kcnj2 gene from sorted mouse hybridoma cells
[0132] The experiment was performed essentially as disclosed in Example 1 with the modification of using primers and probe designed to amplify a single copy of the mouse gene Kcnj2. Primers and probes are as follows:
[0133] Forward primer ctgtcttgccttcgtgctct (SEQ ID NO: 5)
[0134] Reverse primer agcagggctatcaaccaaaa (SEQ ID NO: 6)
[0135] UPL probe (Roche Applied Science catalog number: 04 688 996 001, No. 76 )
[0136] cell number
[0137] As can be seen from the table, even if only one cell was used as starting material, an amplification signal derived from the single copy number mouse gene Kcnj2 could be obtained. In other words, the present invention provides a technical solution for amplifying a single copy gene from a single cell sample.
[0138] Furthermore, it can be observed that the cp value increases if the sample originates from a higher cell number, eg 64 cells. ...
Embodiment 3
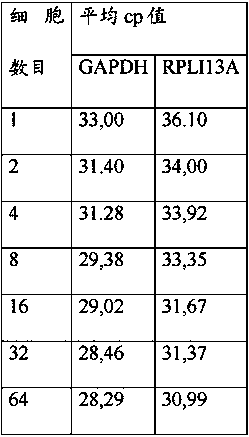
[0142] qPCR for Amplifying the Kcnj2 Gene from Sorted Mouse Hybridoma Cells Using Microtiter Plates Containing Dried Reagents
[0143] The experiment was performed essentially as disclosed in Example 2 with the following changes:
[0144] Fill 10 µl of the solution containing the desired primers and probes into each well of the microtiter plate. The microtiter plate was incubated at 25°C and 200 mBar for 12 hours and then at 25°C and 50 mBar for 4 hours to allow primers and probes to dry on the surface of each reaction well of the microtiter plate.
[0145] Subsequently, cell deposition was performed as follows:
[0146] Columns 1-6 1 cell equivalent / well
[0147] 2 cell equivalents / well in column 7
[0148] 4 cell equivalents / well in column 8
[0149] 8 cell equivalents / well in column 9
[0150] 16 cell equivalents / well in column 10
[0151] 32 cell equivalents / well in column 11
[0152] 64 cell equivalents / well in column 12
[0153] After adding 20 µl of the master m...
PUM
| Property | Measurement | Unit |
|---|---|---|
| volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com