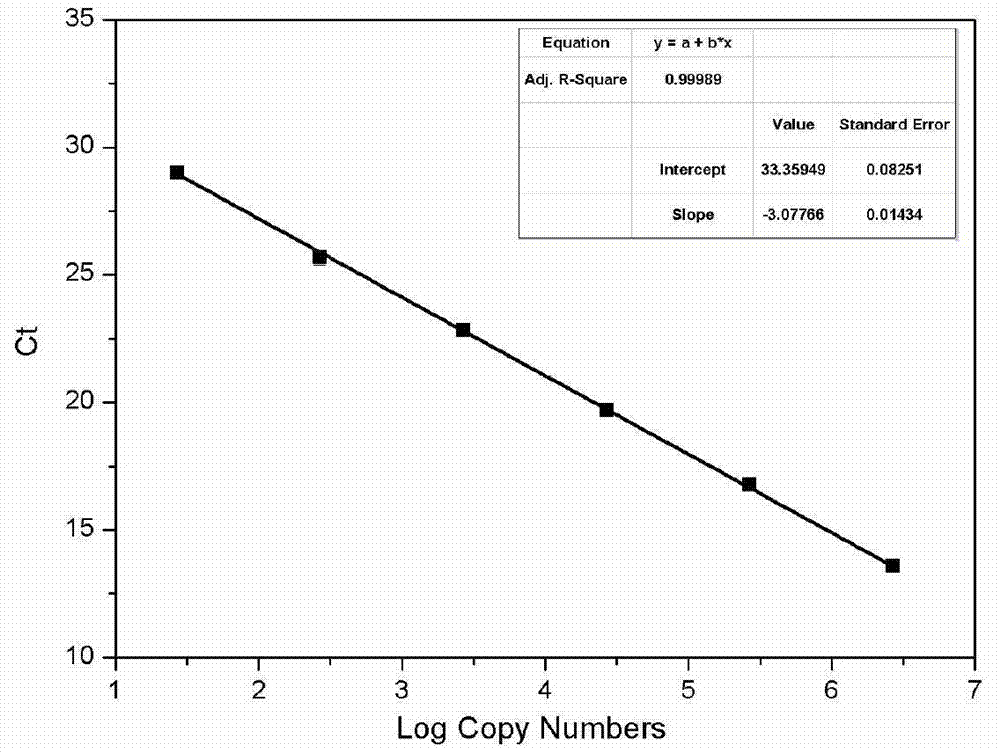
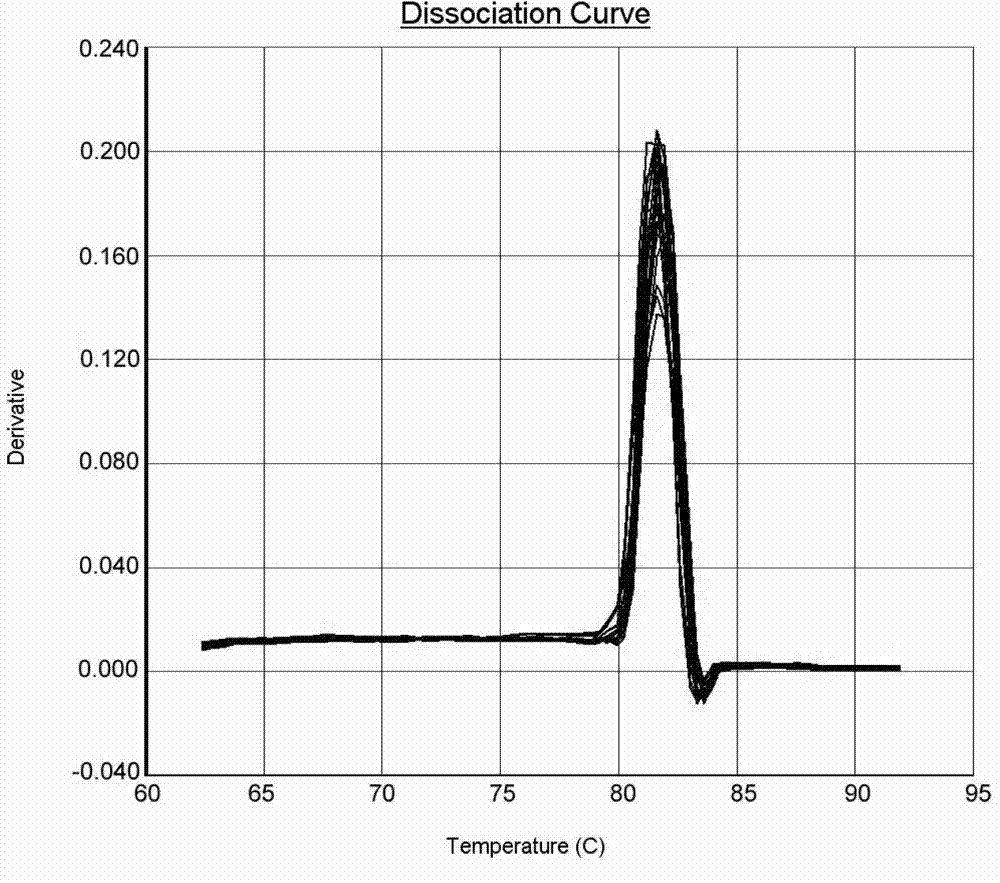
Method for quantitating anaerobic ammonia oxidizing bacteria in sediment of aquiculture environment
A technology for anaerobic ammonium oxidation and aquaculture, applied in biochemical equipment and methods, microbial measurement/inspection, fluorescence/phosphorescence, etc., can solve the problems of low specificity and low diversity of 16SrRNA gene detection, and achieve good results Effect of amplification efficiency and regression relationship
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0029] 1. DNA extraction and quantification
[0030]After collecting aquaculture environmental sediment samples, mix them quickly, put them in dry ice immediately, and transfer them to a -80°C refrigerator for storage. Sediment DNA was extracted using the Fast Prep-24 nucleic acid extractor and the FastDNA Spin kit for soil from MP Biomedicals. The operation method is as follows: add 0.3g of sediment sample (do not exceed 0.3g) to the Lysing MatrixZ tube, and add 1100μl of lysate. Put the above centrifuge tube into the FastPrep-24 nucleic acid extractor, set the speed to 5, and centrifuge for 40s. The centrifuge tube was centrifuged at 14000×g for 10 min at 4°C. Carefully transfer the supernatant to a new 1.5ml centrifuge tube, add 250μl PPS, and gently invert 10 times to mix. Centrifuge the above mixture at 14000×g for 5 min at 4°C, transfer the supernatant into a new centrifuge tube, shake the Binding Matrix Suspension evenly, suspend it, and add 500 μl of Binding Matrix ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com