Glycerol-3- phosphoric desaturase gene relating with glycerol synthesis and uses thereof
A phosphate dehydrogenase and glycerol technology, applied in the application, genetic engineering, plant genetic improvement and other directions, can solve problems such as the decline in economic benefits of glycerol, and achieve the effect of good application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1: Cloning and analysis of the full-length coding regions of DvGPDH1 and DvGPDH2
[0030] Extract total RNA from Salina salina, reverse transcribe the first strand of cDNA, and use the first strand of cDNA as a template to clone into DvGPDH1 by PCR using primers GPDH15'p and GPDH13'p, and clone into DvGPDH2 through primers GPDH25'p and GPDH23'p . In order to further prove the function and application (embodiment three) of these two genes in yeast, by primer GPDH15'p and GPDH25'p, add cloning enzyme point EcoRI and eukaryotic translation conservation sequence Kozak at gene 5' end; Through primer GPDH13 'p and GPDH23'p add clonase site Xho I at the 3' end of the gene.
[0031] The primer sequences are as follows:
[0032] (1) GPDH15'p (introducing EcoR I restriction site and Kozak sequence):
[0033] 5'-AGAATTC AGCATGG TCCTAGGGTCATCACC-3' is the EcoR I recognition site in bold, and the Kozak sequence is underlined;
[0034] (2) GPDH13'p (introduction of Xho ...
Embodiment 2
[0041] Example 2 Protein structure and homology analysis of DvGPDH1 and DvGPDH2
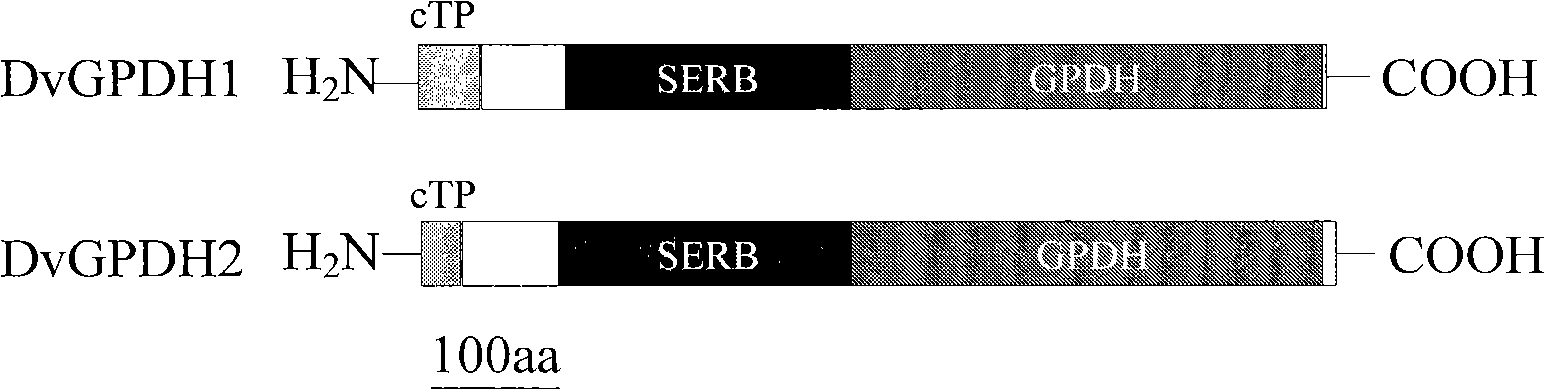
[0042]Using the NCBI (http: / / www.ncbi.nlm.nih.gov / structure / cdd / cdd.shtml) conserved domain analysis system to analyze the proteins encoded by DvGPDH1 and DvGPDH2, the results show that the proteins encoded by the two genes have Unique SERB-GPDH dual domain ( figure 1 ). DvGPDH1 and DvGPDH2 have a GPDH domain of about 400 amino acids at the C-terminus, and compared with the homologous GPDH protein structure in animals and plants, they also have an additional SERB domain of 300 amino acids in length at the N-terminus.
[0043] Homologous analysis of DvGPH1 and DvGPDH2 was carried out by using Vector NTI9.1.0 program. Analysis results show that they have high homology with homologous genes in algae, for example, the homology with Chlamydomonas CrGPDH1 is 45.7% and 45.3%, respectively. The homology with animals or higher plants is relatively low, for example, the homology of yeast ScGpd1p is 19.8...
Embodiment 3
[0045] Example 3 Functional Analysis of DvGPDH1 and DvGPDH2 in Yeast Mutants
[0046] (1) Construction of yeast expression vector
[0047] DvGPDH1 and DvGPDH2 cloned in Example 1 were respectively constructed into yeast expression vector pAJ401 (constitutive strong expression vector) or pYES2 (galactose-inducible expression vector) using the introduced cloning enzyme sites EcoR I and XhoI. In addition, we also constructed vector clones containing only the GPDH domain segment (named DvGPDH1ΔN and DvGPDH2ΔN respectively), and analyzed the function of the gene after the SERB segment was deleted. DvGPDH1ΔN was amplified by primers GPDH1-GPD5’p and GPDH13’p; DvGPDH2ΔN was amplified by primers GPDH2-GPD5’p and GPDH23’p. Use primers GPDH1-GPD5'p and GPDH2-GPD5'p to add cloning enzyme site EcoR I and eukaryotic translation conservation sequence Kozak at the 5' end of the target gene; primers GPDH13'p and GPDH23'p add cloning enzyme at the 3' end of the target gene Point Xho I. The ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
| Isoelectric point | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com