Histone methylated transferase and its encoding gene and application
A technology of histone methylation and transferase, applied in the field of histone methyltransferase and its encoding gene and application, can solve the problem of Arabidopsis dwarf plant, Arabidopsis meristem development retardation, leaves and roots growth retardation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1, the acquisition of histone methyltransferase SDG714
[0037] 1. Discovery of histone methyltransferase SDG714
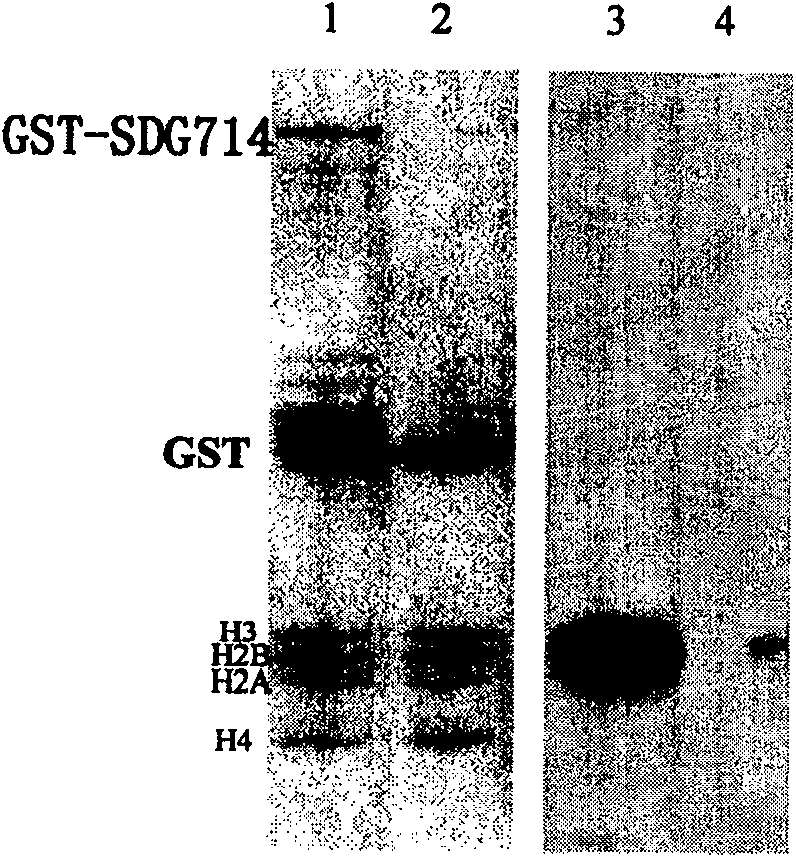
[0038]Data mined through the http: / / www.chrombd.org database showed that there are 38 SET domain proteins in rice, named SDG701 to SDG741. Phylogenetic analysis showed that SDG714 was most similar to KYP / SUVH4 in Arabidopsis, and KYP / SUVH4 belonged to Su(var)3-9 family. SDG714 encodes 663 amino acids (sequence 1 in the sequence listing), contains a conserved SET domain of about 130 amino acids (from the 480th-640th amino acid residue at the amino terminal of sequence 1) and two conserved cysteine moieties body, namely pre-SET (from the 375th to 478th amino acid residue in SEQ ID NO: 1) and post_SET (from the 641st to 663rd amino acid residue in SEQ ID NO: 1). The YDG domain (from the 182nd to the 346th amino acid residue at the amino terminal of SEQ ID NO: 1) is very conserved in the SUVH family of higher plants, but it does not appear in animal...
Embodiment 2
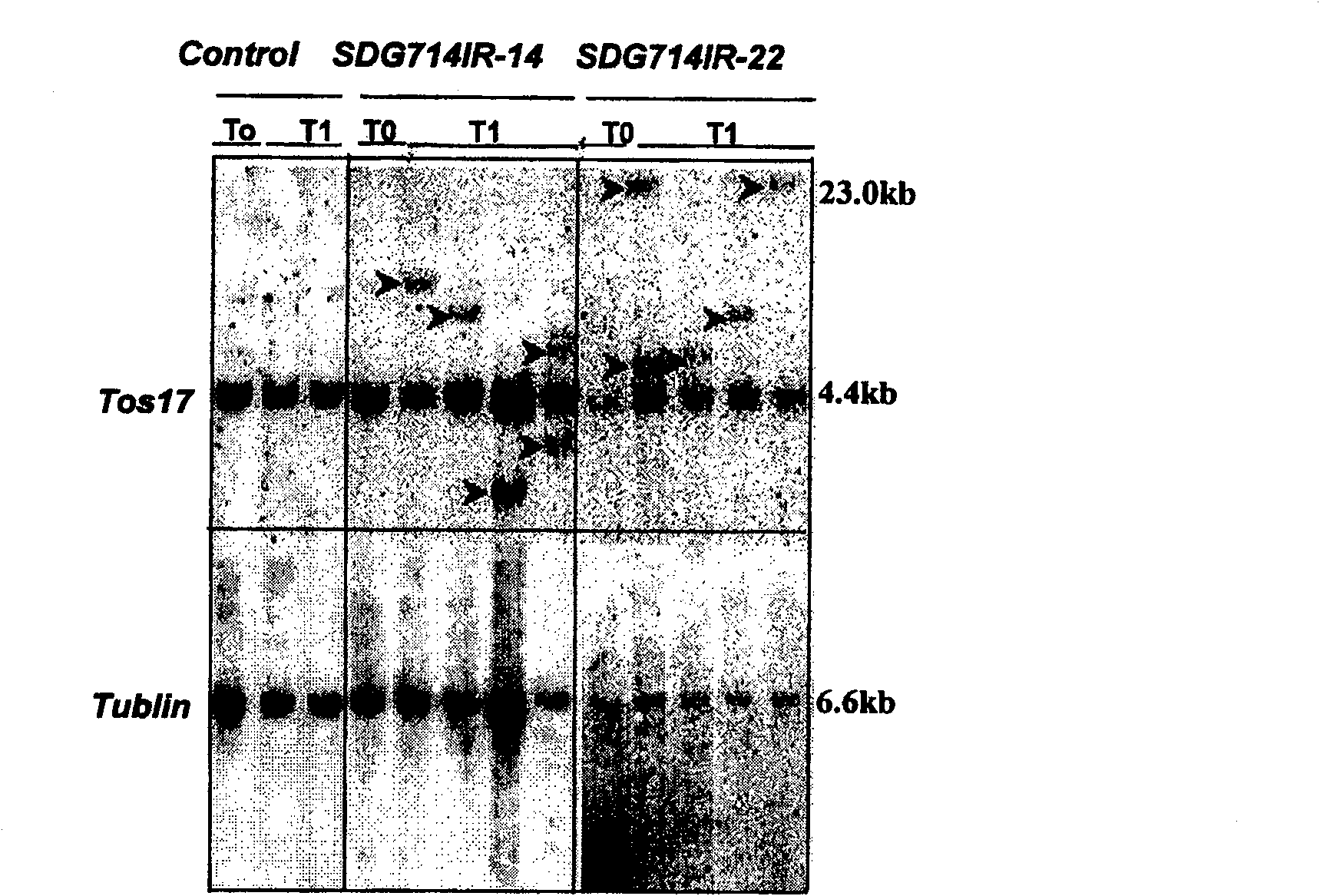
[0050] Example 2, activation of transposition of Tos17 in rice
[0051] 1. Construction of SDG714 RNAi expression vector
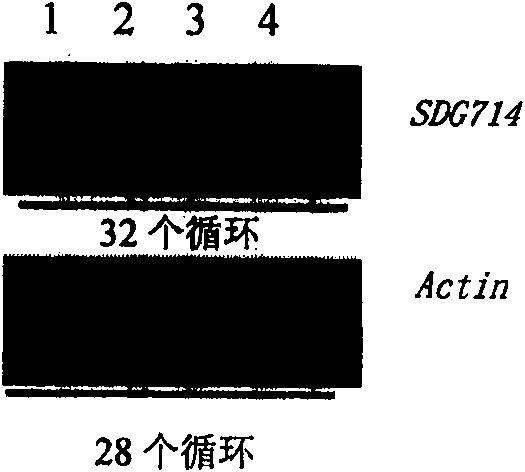
[0052] In order to understand the function of SDG714 in vivo, SDG714 was knocked out by RNAi technology, and the N-terminal of SDG714 and a part of YDG non-conserved part were used as RNAi regions.
[0053] The method of constructing the RNAi expression vector of SDG714 is as follows: extract rice total RNA, use oligo d(T) as a primer, reverse transcribe into cDNA, use primers CX52 (5'-ATTctcgagGCAACTTGTATTGTCATGTCGGG-3') and CX53 (5'-AGCagatctGGGAGCTTCAGCACGAGTAA- 3') PCR amplified deoxyribonucleotides from the 712th to the 1085th position of the 5' end of the sequence 2. After the PCR product was digested with BglII and XbalI, it was inserted into the pUCRNAi vector digested by XhoI / BglII and BamHI / SalI in both forward and reverse directions to obtain the recombinant vector pUC-SDG714RNAi, and then pUC-SDG714RNAi was digested with PstI. The forward and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com