Optical sorting method
a sorting method and optical technology, applied in the field of optical sorting method, can solve the problems of limited scope of the above-mentioned system, limited library size allowed by phage display technology, and inability to direct select activities, etc., to achieve enhanced gene product activity and measure
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
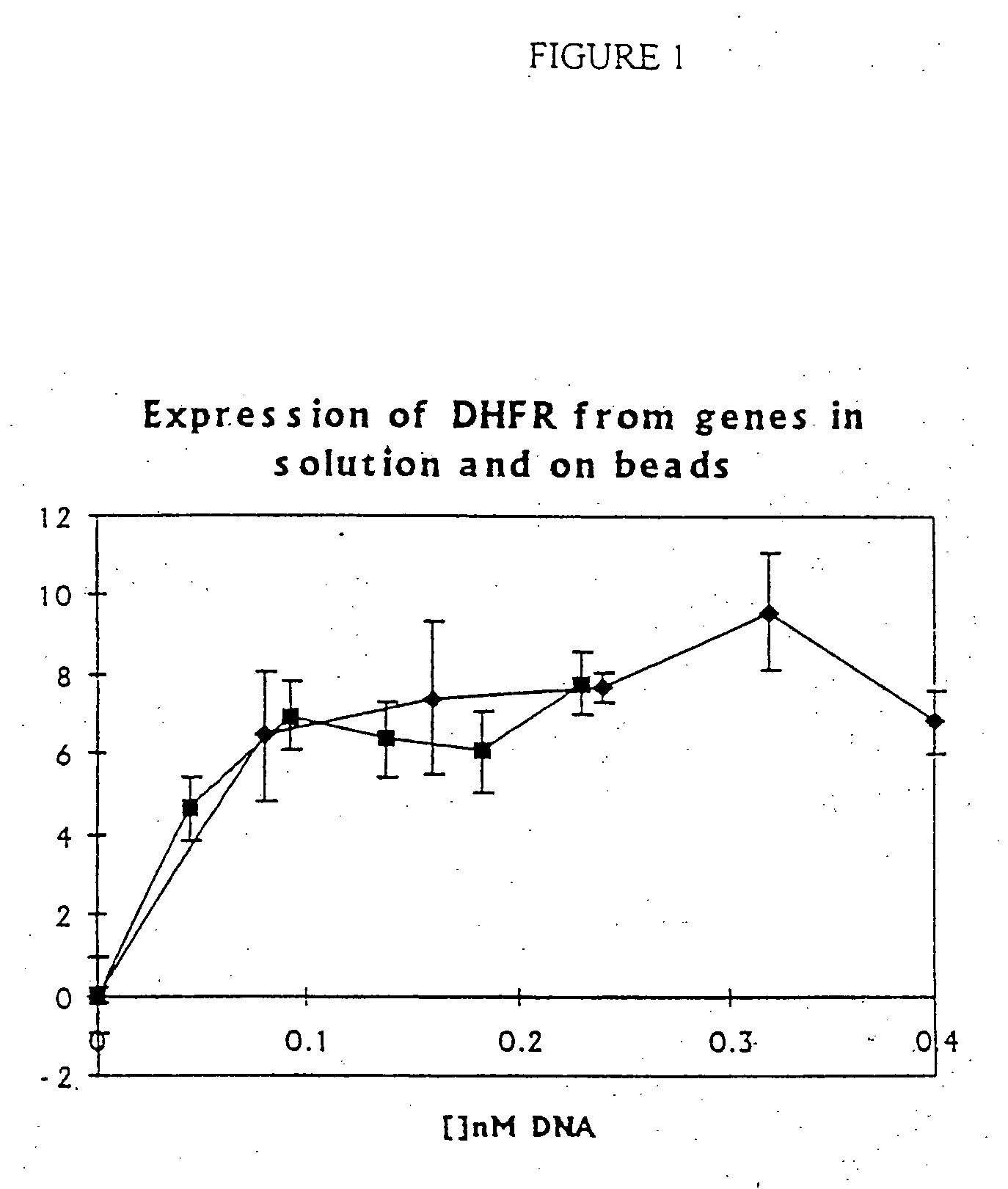
[0216] Enzymes can be expressed from genes in solution and genes attached to paramagnetic microbeads with identical efficiency.
[0217] One format for the selection of genetic elements by using a change in their optical properties is one in which the genetic element comprises a microbead to which the gene is attached. Here it is shown how a gene for an enzyme (E. coli dihydrofolate reductase) can be linked to a paramagnetic bead and is translated in vitro just as efficiently as in solution.
[0218] The E. coli folA gene encoding dihydrofolate reductase (DHFR) is PCR-amplified using oligonucleotides EDHFRFo and EDHFRBa. This DNA is then cloned into the pGEM-4Z vector (Promega) digested with HindIII and KpnI downstream of the lac promoter and the T7 RNA polymerase promoter. The oligonucleotide EDHFRBa appends the efficient phage T7 gene 10 translational start site upstream of the DHFR start codon.
[0219] DNA sequencing identifies a clone which has the correct nucleotide sequence. Bacter...
example 2
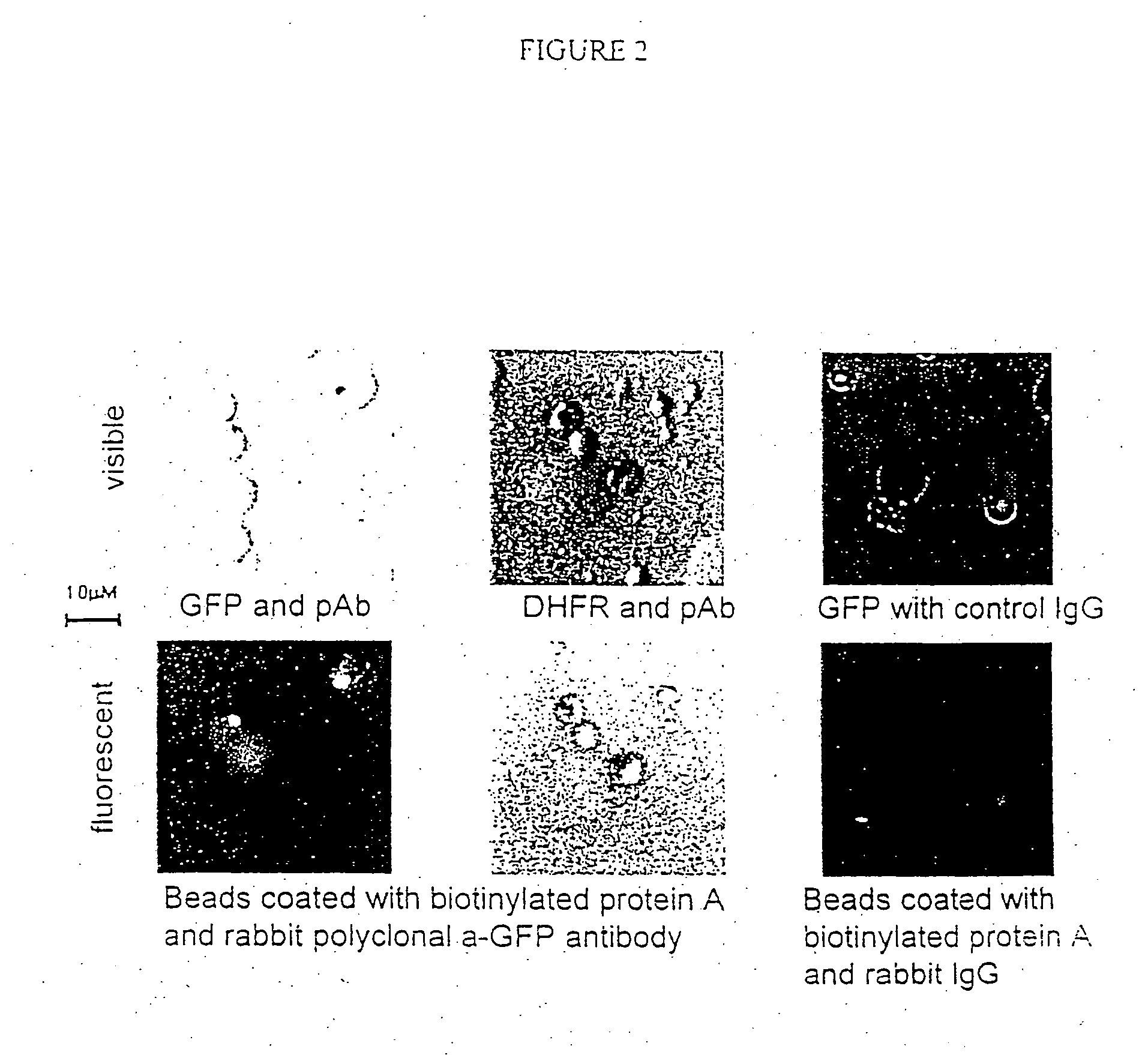
[0226] A fluorescent protein (GFP) can be translated in vitro from genes attached to single microbeads encapsulated in the aqueous compartments of a water-in-oil emulsion and the translated gene-product bound back to the microbeads making them fluorescent.
[0227] One format for the selection of genetic elements is where the genetic element comprises a gene linked to a microbead and the product is coupled back onto the microbead within the microcapsule resulting directly, or indirectly, in a change in the optical properties of the microbead which allows it to be sorted.
[0228] Here it is shown that a fluorescent protein (green fluorescent protein or GFP) can be transcribed and translated in vitro from genes attached to single microbeads encapsulated in the aqueous compartments of a water-in-oil emulsion and the translated gene-product bound back the microbeads making them fluorescent.
[0229] The GFP in pBS / GFP6 plasmid (Siemering et al., 1996) was PCR-amplified using primers GFP-FW a...
example 3
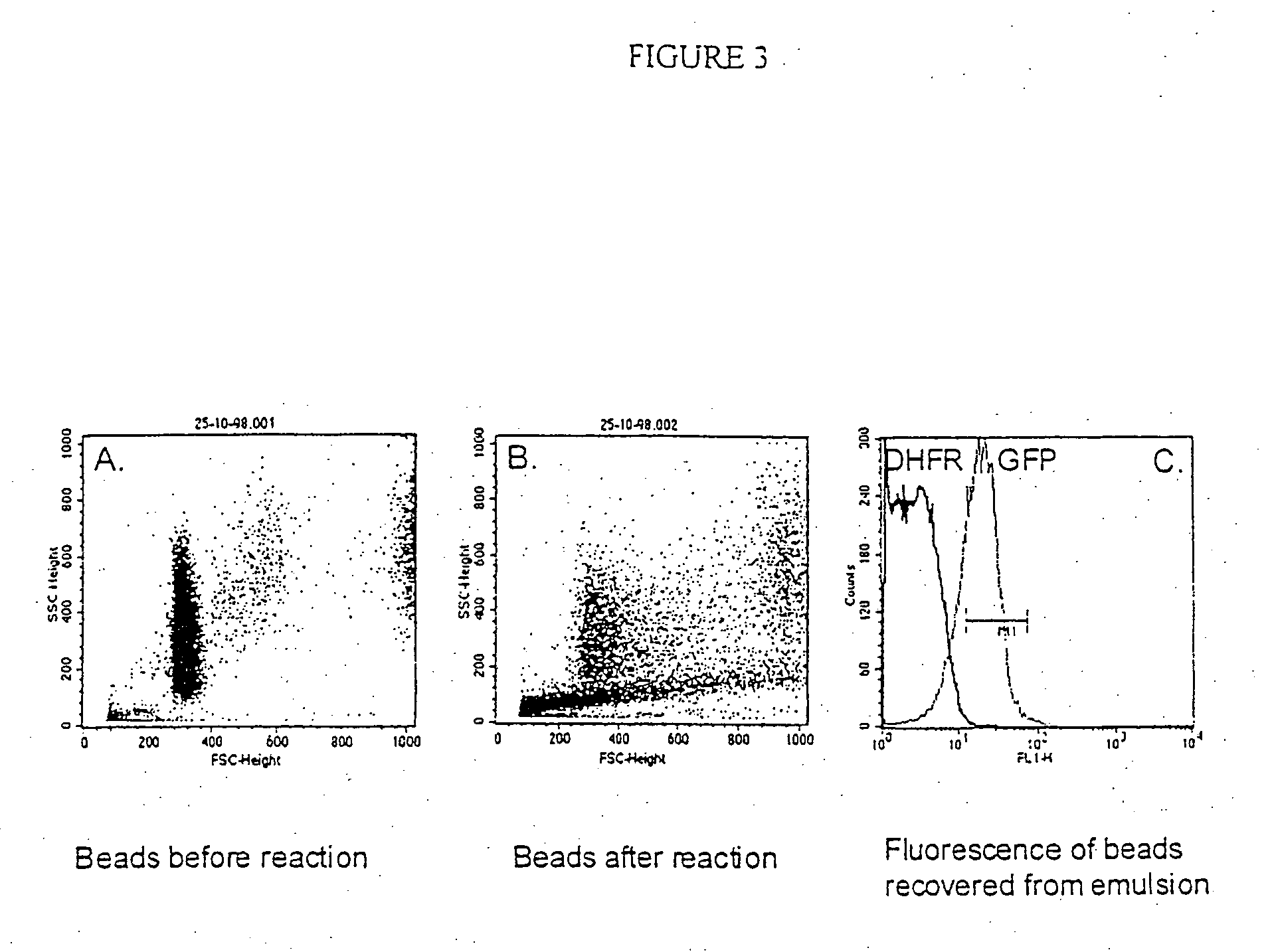
[0235] A fluorescent protein (GFP) can be translated in vitro from genes attached to single microbeads encapsulated in the aqueous compartments of a water-in-oil emulsion, the translated gene-product bound back the microbeads and the increased fluorescence of the microbeads detected by flow cytometry.
[0236] 150 μl streptavidin-coated polystyrene beads (diameter 1 μM; Bangs Laboratories, 2×107 beads / μl) were suspended in 5 mM Tris 7.4 / 1M NaCl / 0.1% Tween20 and split into three aliquots of 50 μl. 0.5 μl of 0.2 μM DNA (T7-folA or T7-GFP) was added to each aliquot of beads, incubated at 43° C. for 15 min, washed three times in 25 mM NaH2PO4, 125 mM NaCl, 0.1% Tween20, pH 7.0 (PBS / 0.1% Tween20), resuspended in 40 μl TBST and 10 μl 80 μM biotinylated protein A (Sigma) was added (to give final concentration of 15 μM). After incubation for 30 minutes at room temperature, the beads were washed three times in PBS / 0.1% Tween20 and resuspended in 20 μl 1:10 dilution rabbit anti-GFP polyclonal a...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com